Amin Katouzian

|
Dr. Amin Katouzian Senior Research Scientist Chair for Computer Aided Medical Procedures & Augmented Reality Fakultät für Informatik Technische Universität München Boltzmannstr. 3 85748 Garching b. München Room: MI 03.13.053 Phone: +49 89 289 17081 Fax: +49 89 289 17059 E-Mail: Skype ID: Amin.Katouzian |
Education
- Ph.D. in Biomedical Engineering, Columbia University, New York, USA
- M.Sc. in Electrical Engineering, Fairleigh Dickinson University, New Jersey, USA
- B.Sc. in Electrical Engineering, Sistan & Baluchestan University, IRAN
Awards
- Samsung GRO Award of $100,000 per year up to three years.
- Volcano Corporation (Rancho Cordova, CA) Fellowship, $78,100, Sep. 2008-Sep. 2010.
- Siemens (Princeton, NJ) Fellowship, $50,000, Jun. 2007-Dec. 2007.
- Boston Scientific (Fremont, CA) Fellowship, $122,500, Jan. 2006-Jun. 2007.
- Colonel Fairleigh S. Dickinson (Teaneck, NJ) Scholarship, $7,500, Jan. 2002-Sep. 2003.
- Third place in North New Jersey IEEE student paper presentation contest, graduate category, Teaneck, NJ, March 2003.
Research Interests
- Computer Aided Surgery and Diagnosis.
- Augmented Reality in Medical Procedures.
- Computer Vision for Medical Applications.
- Telemedicine.
- Multidimensional Cue Extraction and Pattern Recognition.
- Reconstruction, Registration, Visualization, and Segmentation of 2-4D Images.
- Multiresolution Analysis.
- Speech Recognition and Perception.
Active Research Projects
Estimation of Flow Parameters from X-ray DataThis project focuses on the quantification of blood flow based on 2D and 3D angiographic X-ray image data, which is of particular interest during minimally invasive interventional procedures such as recanalizations of stenotic vessels and embolizations of arteries that feed tumors or vascular malformations in the patient's brain, for example. Our projects cover the evaluation of model-based approaches involving CFD (computational fluid dynamics) simulations as well as methods that aim at estimating the optical flow in the images and deriving quantitative blood flow information from these results. |
Big Data Analysis for Medical ApplicationsWidespread use of electronic health records (EHR) led to vast amount of medical data being collected. The data is characterized by large sample size, heterogeneity of variables, unstructured information, missing information, and time-dependent variables, to name a few. These facts render commonly used statistical tools to perform analysis inadequate and require the development of sophisticated algorithms to overcome these challenges. For instance, the set of variables recorded for each patient can naturally be decomposed into groups, known as views. Most machine learning algorithms ignore this multi-view relationship. Instead, they can either be trained on each view separately or on a concatenation of all views to form a single view. Considering these relationships recently came to attention of the research community that proposed co-training, multiple kernel learning and subspace learning to address this problem. Although existing methods show promising results in their respective tasks, it is difficult to apply them to real-world clinical data, where highly heterogeneous features (continuous, categorial, ordinal) and missing values are common, because not all tests can be performed on all patients. As a result, multi-view learning algorithms have rarely been applied to medical problems. |
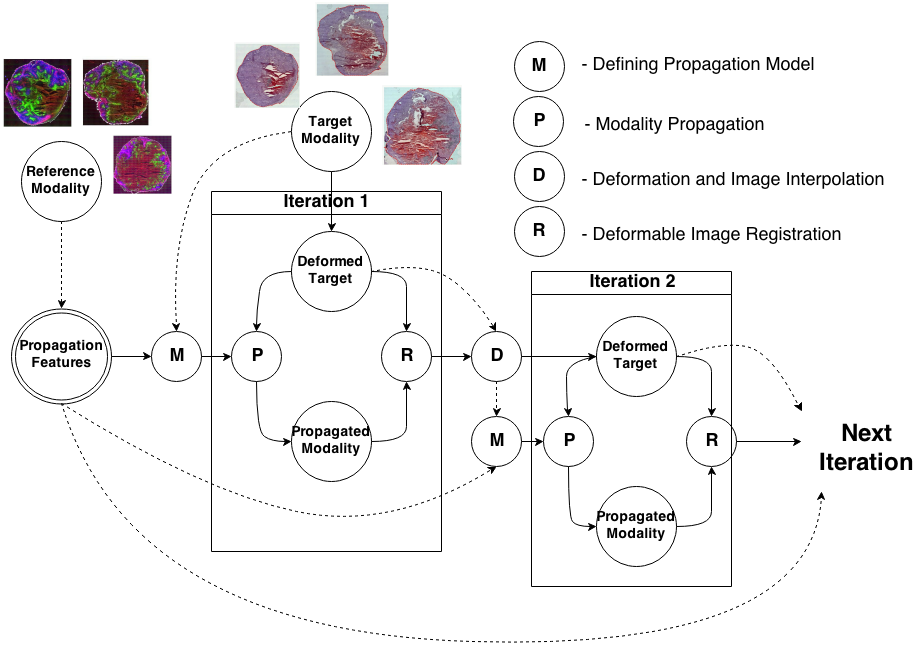
Knowledge Propagation Models for Image RegistrationTo register modalities with complex intensity relationships, we leverage machine learning algorithm to cast it into a mono modal registration problem. This is done by extracting tissue specific features for propagating anatomical/structural knowledge from one modalitiy to an other through an online learnt propagation model. The registration and propagation steps are iteratively performed and refined. For proof-of-concept, we employ it for registering (1) Immunofluorescence to Histology images and (2) Intravascular Ultrasound to Histology Images. |
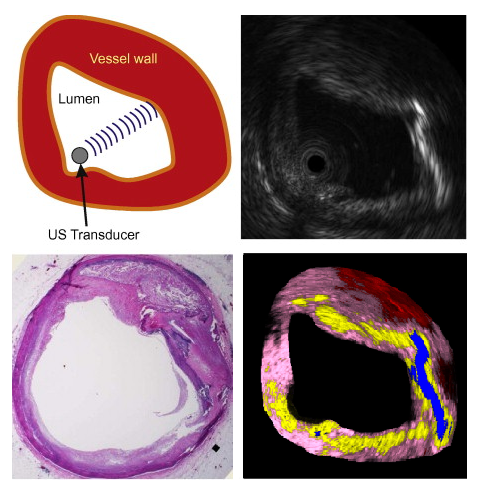
OCT Tissue ClassificationOptical coherence tomography (OCT), employing light rather than ultrasound, is a high-resolution imaging technology that permits a precise assessment of biological tissue. Used intravascular, OCT is increasingly used for assessing safety and efficacy of intracoronary devices, such as drug-eluting stents and bioabsorbable stents. Obtained images provide insights regarding stent malposition, overlap, and neointimal thickening, among others. Recent OCT histopathology correlation studies have shown that OCT can be used to identify plaque composition, and hence it is possible to distinguish “normal” from “abnormal” neointimal tissue based on its visual appearance. The aim of this project is to develop a novel method for the automatic analysis of tissue in IVOCT. Automatic tissue classification will allow for a quantitative and potentially more time-efficient and objective analysis of IVOCT data. For instance, classifying neointimal tissue as either “mature” or “immature” can be used to assess the disease state of patients; as a potential predictor of late stent-failure events such as stent thrombosis and restenosis. |
Ultrasound Based Tissue Characterizationclinicians are challenged when colocated heterogeneous tissue backscatter mixed signals appearing as non-unique intensity patterns in B-mode ultrasound image. Tissue characterization algorithms have been developed to assist clinicians to identify such heterogeneous tissues and assess lesion stage. We propose a novel technique coined as Stochastic Driven Histology (SDH) that is able to provide information about co-located heterogeneous tissues. It employs learning of tissue specific ultrasonic backscattering statistical physics and signal confidence primal from labeled data for predicting heterogeneous tissue composition in plaques. We employ a random forest for the purpose of learning such a primal using sparsely labeled and noisy samples. In clinical deployment, the posterior prediction of different lesions constituting the plaque is estimated. |
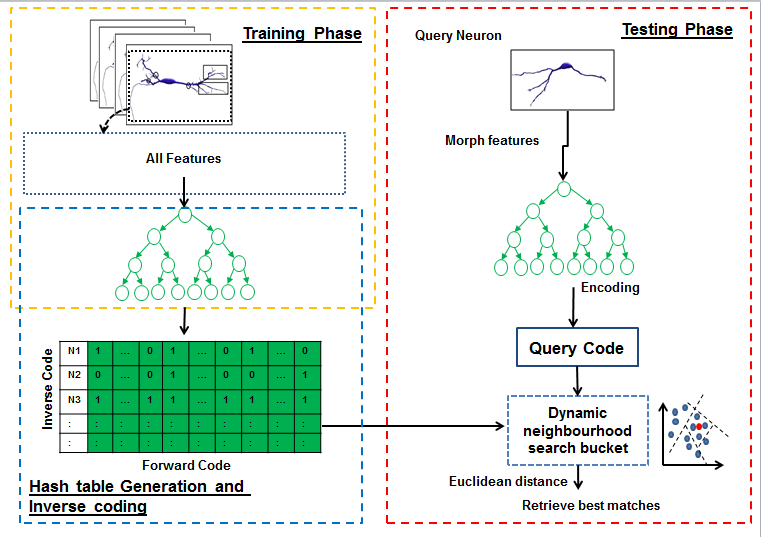
Hashing forests for morphological search and retrieval in neuroscientific image databasesIn this project, for the first time, we propose a data-driven search and retrieval (hashing) technique for large neuron image databases. The presented method is established upon hashing forests, where multiple unsupervised random trees are used to encode neurons by parsing the neuromorphological feature space into balanced subspaces. We introduce an inverse coding formulation for retrieval of relevant neurons to effectively mitigate the need for pairwise comparisons across the database. Experimental validations show the superiority of our proposed technique over the state-of-the art methods, in terms of recall for a particular code size. This demonstrates the potential of this approach for effective morphology preserving encoding and retrieval in large neuron databases. |
Student Projects
- Implementation and development of a new service to report, analyze, visualize neurons for neuroscientists in Python (IDP, Finished).
- Cardiac Magnetic Resonance Image Analysis and Modeling for Patients with Implantable Cardioverter Defibrillators (ICDs) (Master Thesis, Finished).
- Integration of a WiFi? Interface and Win7 C#-GUI development for control and surveillance of a Heart Assist Device in animal experiments (IDP, Finished).
- Knowledge propagation models for deformable image registration (Master Thesis, Finished).
- Implementation and development of wavelet-packet-based tissue characterization for IVUS image in C++ and MeVisLab? (IDP, Finished).
Reviewer
- Journal of Medical Image Analysis.
- Journal of IEEE Transaction on Medical Imaging.
- Journal of IEEE Transaction on Biomedical Engineering.
- Journal of IEEE Transaction on Information Technology in Biomedicine.
- Journal of Digital Signal Processing.
- Journal of Computers in Biology and Medicine.
- Journal of Computerized Medical Imaging and Graphics
- Journal of Cardiovascular Imaging.
- Journal of Cardiovascular Magnetic Resonance.
- Medical Image Computing and Computer Assisted Intervention (MICCAI) 2013.
- Design of Medical Devices Conference, Europe 2013.
- International Symposium on Mixed and Augmented Reality (ISMAR) 2012.
- Medical Image Computing and Computer Assisted Intervention (MICCAI) 2012.
- 15th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)-CVII.
Editorial Activity
- Program Committee: International Conference on Systems in Medicine and Biology (ICSMB), Kharagpur, India, 2016.
- Steering Committee: the 18th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)-CVII/STENT workshop, Munich, Germany, 2015.
- Program Committee, the MICCAI workshop on Computing and Visualization for Intravascular Imaging and Computer Assisted Stenting MICCAI-CVII-STENT, Boston, USA, 2014.
- Executive Committee, the 4th International Conference on Information Processing in Computer-Assisted Interventions (IPCAI), Heidelberg, Germany, 2013.
- Program Committee, the 15th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)-CVII, Toronto, Canada, 2011.
Invited Talks
- PicoSEC?-MCNet Training Workshop, “The necessity of personalized monitoring for patients with cardiovascular disease,” Munich, March 19th 2015.
- Otto-von-Guericke University (Magdeburg, Germany), Computer Science Department, “Computer-assisted decision support for coronary atherosclerosis diagnosis and treatment,” February 5th, 2015.
- Tehran University, Electrical Engineering and Computer Science Department, “Computer-assisted decision support for coronary atherosclerosis diagnosis and treatment,” January 25th, 2015.
- MICCAI-CVII workshop, "The missing link in healthcare system; personalized monitoring," MIT Boston, 2014.
- University of Barcelona, Computer Vision Center, "patient-Specific Translational research on Atherosclerosis and Diagnosis (STAnD?)@CAMP@TUM". February 22nd, 2013.
- PicoSEC?-EndoTOFPET-US workshop on intraoperative imaging and navigation solutions, January 16th, 2013.
- Technical University of Munich, Department of Mathematics, "Applications of multiscale analysis in quantification of atherosclerosis disease," November 10th, 2011.
- Sabanci University (Istanbul, Turkey), Department of Engineering and Natural Sciences, "Quantifying Atherosclerosis: IVUS Imaging For Lumen Border Detection and Plaque Characterization," December 22nd, 2010.
- Volcano Corporation (Sacramento, CA), "Detection of Lumen Border in IVUS images via Three-Dimensional Brushlet Analysis," June 28th, 2009.
Patent
- Ali Kamen, Lance Ladic, Sebastian Pölsterl, Adnan Kastrati, Amin Katouzian, Nassir Navab, “A novel cost-effective method for stratification of CAD patients for revascularization procedure based on minimizing the adverse effect,” 2013.
- Debdoot Sheet, Amrita Chaudhary, Jyotirmoy Chatterjee, Ajoy Kumar Ray, Amin Katouzian, "Methods and System for Characterizing Tissues in Optical Coherence Tomography," 2013.
- Amin Katouzian, Nassir Navab, “Intelligent Implanted Health Sensing Device and Assembly”, 2012.
- Amin Katouzian, Andrew F. Laine, Debdoot Sheet, Athanasios Karamalis, Abouzar Eslami, Stephane G. Carlier, Nassir Navab, “System and Method for Characterizing Tissues in Intravascular Ultrasound using Statistical Physics," 2012.
- Amin Katouzian, Elsa D. Angelini, Auranuch Lorsakul, Bernhard Sturm, Andrew F. Laine, “Denoising and Lumen Border Detection of Intravascular Ultrasound via Segmentation of Directional Wavelet Representations,” Serial No. 61/200,987, December 5, 2008.
- Amin Katouzian, Babak Baseri, Elisa E. Konofagou, Andrew F. Laine, “Systems and Methods for Intravascular Tissue Characterization and Border Detection,” WO2009023626, 2009.
- Amin Katouzian, Elisa E. Konofagou, Andrew F. Laine, “Unsupervised Texture-Derived Atherosclerotic Plaque Characterization Algorithm through Backscattered Intravascular Ultrasound Signals,” Serial No. 60/991,508, November 30, 2007.
- Amin Katouzian, Elisa E. Konofagou, Andrew F. Laine, “Intravascular Ultrasound (IVUS) Tissue Characterization and Border Detection through Backscattered Signals by Wavelet Packet Analysis,” Serial No. 60/955,249, August 10, 2007.
Publication
| UsersForm | |
|---|---|
| Title: | Dr. |
| Circumference of your head (in cm): | |
| Firstname: | Amin |
| Middlename: | |
| Lastname: | Katouzian |
| Picture: | |
| Birthday: | |
| Nationality: | Iran |
| Languages: | |
| Groups: | |
| Expertise: | |
| Position: | Scientific Staff |
| Status: | Alumni |
| Emailbefore: | amin.katouzian |
| Emailafter: | cs.tum.edu |
| Room: | MI 03.13.053 |
| Telephone: | +49 89 289 17081 |
| Alumniactivity: | |
| Defensedate: | |
| Thesistitle: | |
| Alumnihomepage: | |
| Personalvideo01: | |
| Personalvideotext01: | |
| Personalvideopreview01: | |
| Personalvideo02: | |
| Personalvideotext02: | |
| Personalvideopreview02: | |