Ing. John T LaMaster, M.Sc.

John LaMaster
PhD Candidate
- Groups: Image-Based Biomedical Modeling Group (IBBM)
- Supervisors: Prof. Dr. Björn Menze and Prof. Dr. med. Jan Stefan Kirschke
- Room: MSB/IMETUM 1.101
- Building: Munich School of Bioengineering (MSB) Boltzmannstr. 11, Garching Forschungszentrum (map)
- Email:
About me
I studied Biomedical Engineering in St. Louis, MO (USA) at Saint Louis University (B.S. 2014) where I focused on tissue engineering. I then completed the Erasmus Mundus' Common European Master's Course in Biomedical Engineering (CEMACUBE) at RWTH Aachen University (RWTH Aachen, Germany) and the Czech Technical University in Prague (ČVUT Prague, Czech Republic) where I specialized in Medical Imaging Instrumentation (M.Sc. 2016). My interests have evolved from regenerative medicine in my bachelor's to medical imaging in the master's, and I am now focusing on medical image processing and analysis based on deep learning techniques primarily using MR images, MR spectroscopy, and tumor simulations. My current research, funded by the German Academic Exchange Service (DAAD), focuses on developing deep learning techniques for automating the processing and analysis of MRSI data which will eventually lead to computer-aided diagnostic (CAD) devices for use in characterizing and diagnosing brain tumors.Background
- 2018 - present Ph.D. Technical University of Munich, Munich, Germany - DAAD Fellow
- 2016 M.Sc. Biomedical Engineering - Czech Technical University in Prague, Prague, Czech Republic
- 2014 - 2015 RWTH Aachen, Aachen, Germany
- 2014 B.Sc. Biomedical Engineering - Saint Louis University, St. Louis, MO USA
- 2011 Study Abroad - Saint Louis University - Madrid, Madrid, Spain
Research Interests
- Studying the domain shift of DL models trained on synthetic MRSI data
- Autormating the preparation and quantification of large-scale MRSI brain spectra
- Multi-modal disease classification of brain tumors using MR data
- Simulating realistic MRI data
What is Magnetic Resonance Spectroscopy?
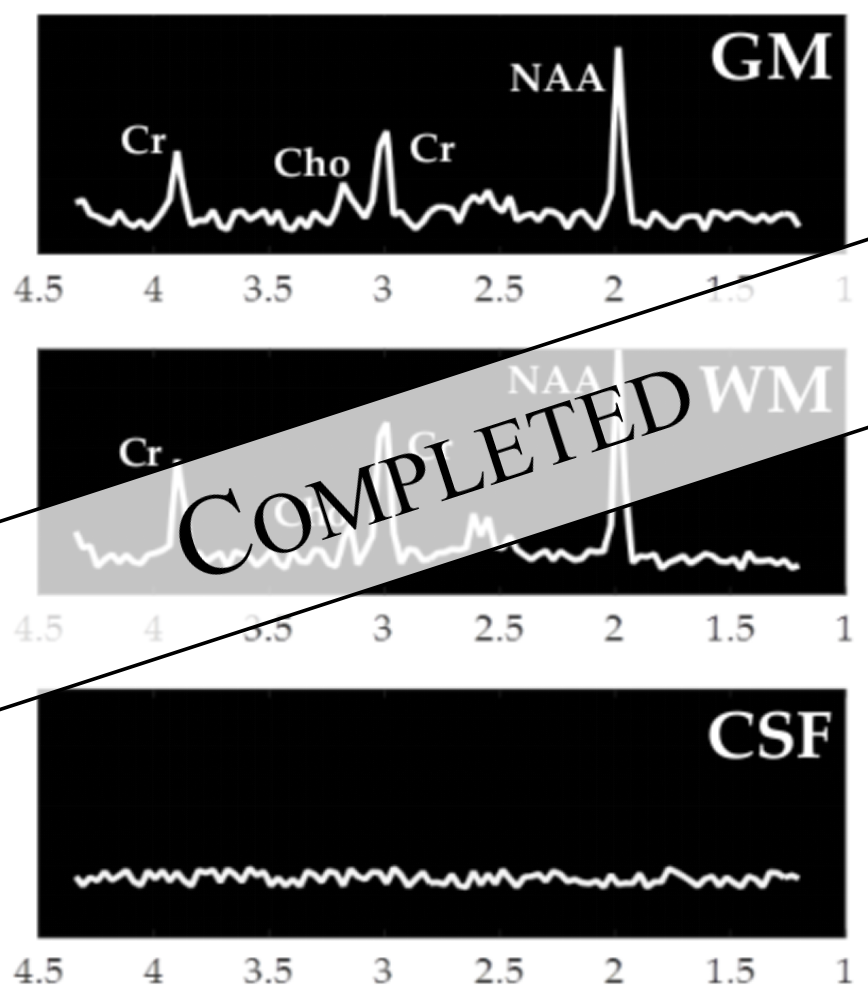
MRS is a quantitative MR technique used to characterize the metabolism or metabolic composition of tissue in vivo. This is done by analyzing the specific frequencies present in the FID, instead of just considering the magnitude which is used for qualitative imaging. These frequencies are metabolite dependent, making them easy to identify. While all protons resonate at the same Larmor frequency, the precessions that occur when excited by the RF pulse cause small changes in the magnetic field of the nucleus. MR-visible molecules are visible because there are many of these small perturbations that cumulatively change the local effective magnetic field of the molecule. This change in the magnetic field changes the molecule's resonant frequency. In the frequency domain, this change in resonant frequency shifts the frequency peak to the left or the right. This chemical shift is unique to each molecule and by comparing peaks of different metabolites, maps of relative metabolite concentrations can be generated. This type of information is useful for characterizing tissue, including potential tumors.What is Deep Learning?
Deep Learning is an off-shoot of machine learning that focuses on using models with multiple hidden layers between the input and output layers. During training, data is fed into the model where many mathematical operations occur until the output is calculated. Then based on chosen optimizaiton scheme, a loss or penalty is calculated depending on how good or bad the output is. Some form of gradient descent is used to update the parameters of the model that used in the mathematical operations mentioned above. The model continues to update the parameters until the model converges. This basic framework can be used for things such as object detection, image classification, multivariate regression, reinforcement learning, image and text translations, and many more. As different as these applications may seem, they all have at least one thing in common which is the fundamental task of all machine learning techniques: pattern recognition. Networks can easily become very complicated in order to acheive a specific goal. The way these networks are optimized can also vary greatly. But the underlying goal of every model is to learn patterns that lead to accuract predictions, classifications, and regressions.Current Collaborations
- Sara J. Nelson Lab, Center for Inteligent Imaging, University of California, San Francisco (UCSF)
- Multimodal Metabolic Brain Imaging Lab, Center for Inteligent Imaging, University of California, San Francisco (UCSF)
- Body Magnetic Resonance Research Group, Klinikum Rechts der Isar and Technical University of Munich (TUM)
Current Research Projects
- Improving Generalizability of Deep Learning Models for MRS Quantificaiton of Brain Metabolites
- Unsupervised Approaches to MRS Quantification of Brain Tumor Spectra
- Improving Quantification of Fat- and Water-Fractions of Intervertebral Bone Marrow MR Spectroscopy
Available Projects
Please contact me for available student projects (M.Sc thesis, B.Sc. thesis, semester projects) if you are interested in medical image analysis for brain applications based on MRI images and MR spectroscopy. Please include your CV, transcript, and letter of motivation.Improving Generalizabilty through Generative Adversarial Domain Adaptation in MR Spectroscopy
Project Page
Transformer-Based Regression Model for Metabolite Quantification in MR Spectroscopic Imaging

Project Page
Deep Learning Approach for Automating Quality Assessment of MR Spectroscopic Imaging Data

Coming Soon
Deep Learning Approach for Water-Fat Quantification of Bone Marrow MR Spectroscopy

Coming Soon
Image credits: Dr. Dhritiman Das; None; Dr. Roland Kreis; None; None
Former Students
- Linus Kreitner (B.Sc Thesis, 2021) (Thesis) (Repository)
 Google Scholar
Google Scholar orcid.org/0000-0002-2149-771X
orcid.org/0000-0002-2149-771X
