Sailesh Conjeti
|
|
Sailesh Conjeti Currently: Post-Doc, German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany. Past: PhD Student Chair for Computer Aided Medical Procedures & Augmented Reality Fakultät für Informatik Technische Universität München Boltzmannstr. 3 85748 Garching b. München Room: MI 03.13.053 Phone: +49 89 289 17081 Fax: +49 89 289 17059 E-Mail: sailesh.conjeti@tum.de ; sailesh.conjeti@dzne.de Skype ID: saileshconjeti |
Education
- Master of Technology (2014) in Medical Imaging and Informatics, Indian Institute of Technology Kharagpur, INDIA
- Bachelor of Engineering - with honours (2012) in Electrical and Electronics Engineering, Birla Institute of Technology and Science Pilani, INDIA
Awards
- Student Travel Grant for MICCAI 2017 (co-authored paper).
- Elsevier Medical Image Analysis Best Paper Award, MICCAI 2016 for paper on Metric Hashing Forests (First Author).
- Young Scientist Award - Runner Up in MICCAI 2015 (First Author).
- Institute Silver Medal from Indian Institute of Technology Kharagpur, for best academic performance in Post Graduate Course in Medical Imaging and Informatics - July 2014
- Deutscher Akademischer Austauschdienst (DAAD) (Bonn, GERMANY) Scholarship, Sep. 2013 - Mar. 2014 - Awarded to pursue Master's Thesis at Chair for Computer Aided Medical Procedures & Augmented Reality, Fakultät für Informatik, Technische Universität München.
- Ministry of Human Resources and Development, Government of India Scholarship for pursuing graduate studies in Medical Imaging and Informatics after qualifying Graduate Aptitude Test in Engineering. (Percentile:99.81 % among 1,10,125 candidates).
- Best Outgoing Student of the Year 2012 awarded by the Department of of Electrical and Electronics Engineering, BITS Pilani for Overall Excellence.
- BITSAA IRU Research Travel Grant, 2012 for attending the IEEE EMBS BHI 2012, Shenzhen, China.
- National Talent Search(NTS) Scholarship by National Council for Education, Research and Training(NCERT) for scholastic excellence in 2007.
Active Research Projects
Big Data Analysis for Medical ApplicationsWidespread use of electronic health records (EHR) led to vast amount of medical data being collected. The data is characterized by large sample size, heterogeneity of variables, unstructured information, missing information, and time-dependent variables, to name a few. These facts render commonly used statistical tools to perform analysis inadequate and require the development of sophisticated algorithms to overcome these challenges. For instance, the set of variables recorded for each patient can naturally be decomposed into groups, known as views. Most machine learning algorithms ignore this multi-view relationship. Instead, they can either be trained on each view separately or on a concatenation of all views to form a single view. Considering these relationships recently came to attention of the research community that proposed co-training, multiple kernel learning and subspace learning to address this problem. Although existing methods show promising results in their respective tasks, it is difficult to apply them to real-world clinical data, where highly heterogeneous features (continuous, categorial, ordinal) and missing values are common, because not all tests can be performed on all patients. As a result, multi-view learning algorithms have rarely been applied to medical problems. |
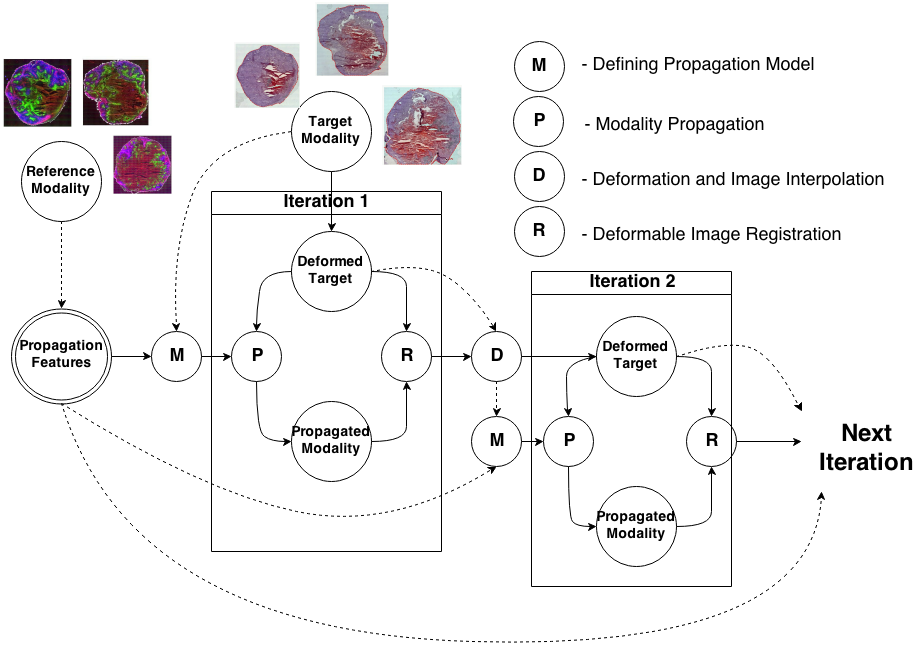
Knowledge Propagation Models for Image RegistrationTo register modalities with complex intensity relationships, we leverage machine learning algorithm to cast it into a mono modal registration problem. This is done by extracting tissue specific features for propagating anatomical/structural knowledge from one modalitiy to an other through an online learnt propagation model. The registration and propagation steps are iteratively performed and refined. For proof-of-concept, we employ it for registering (1) Immunofluorescence to Histology images and (2) Intravascular Ultrasound to Histology Images. |
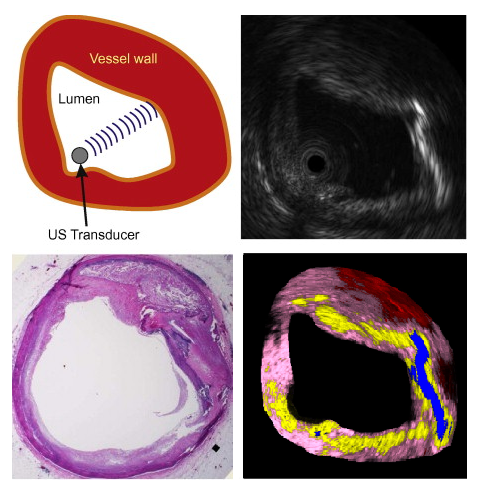
OCT Tissue ClassificationOptical coherence tomography (OCT), employing light rather than ultrasound, is a high-resolution imaging technology that permits a precise assessment of biological tissue. Used intravascular, OCT is increasingly used for assessing safety and efficacy of intracoronary devices, such as drug-eluting stents and bioabsorbable stents. Obtained images provide insights regarding stent malposition, overlap, and neointimal thickening, among others. Recent OCT histopathology correlation studies have shown that OCT can be used to identify plaque composition, and hence it is possible to distinguish “normal” from “abnormal” neointimal tissue based on its visual appearance. The aim of this project is to develop a novel method for the automatic analysis of tissue in IVOCT. Automatic tissue classification will allow for a quantitative and potentially more time-efficient and objective analysis of IVOCT data. For instance, classifying neointimal tissue as either “mature” or “immature” can be used to assess the disease state of patients; as a potential predictor of late stent-failure events such as stent thrombosis and restenosis. |
Ultrasound Based Tissue Characterizationclinicians are challenged when colocated heterogeneous tissue backscatter mixed signals appearing as non-unique intensity patterns in B-mode ultrasound image. Tissue characterization algorithms have been developed to assist clinicians to identify such heterogeneous tissues and assess lesion stage. We propose a novel technique coined as Stochastic Driven Histology (SDH) that is able to provide information about co-located heterogeneous tissues. It employs learning of tissue specific ultrasonic backscattering statistical physics and signal confidence primal from labeled data for predicting heterogeneous tissue composition in plaques. We employ a random forest for the purpose of learning such a primal using sparsely labeled and noisy samples. In clinical deployment, the posterior prediction of different lesions constituting the plaque is estimated. |
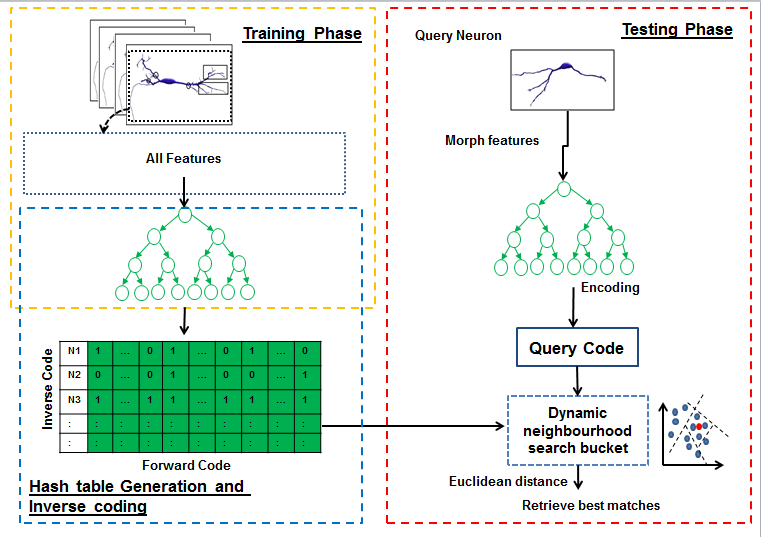
Hashing forests for morphological search and retrieval in neuroscientific image databasesIn this project, for the first time, we propose a data-driven search and retrieval (hashing) technique for large neuron image databases. The presented method is established upon hashing forests, where multiple unsupervised random trees are used to encode neurons by parsing the neuromorphological feature space into balanced subspaces. We introduce an inverse coding formulation for retrieval of relevant neurons to effectively mitigate the need for pairwise comparisons across the database. Experimental validations show the superiority of our proposed technique over the state-of-the art methods, in terms of recall for a particular code size. This demonstrates the potential of this approach for effective morphology preserving encoding and retrieval in large neuron databases. |
Research Interests
* Machine Learning: Deep Learning, Image Retrieval, Segmentation, Domain Adaptation, Adversarial Learning. * Modalities: Histology, X-Ray, Ultrasound, MR.Professional Associations and Memberships
- MICCAI Society Student Member
- IEEE Student Member
Reviewer
- Medical Image Analysis
- Biomedical Optics Express
- IEEE Transactions on Medical Imaging.
- IEEE Transactions on Intelligent Transportation Systems.
- Medical and Biological Engineering and Computing.
- Computers in Biology and Medicine.
Student Projects
Contact me.Resume
Click here to download the latest resume (last updated: Sept 2015)Publications
| 2019 | |
| F. Navarro, S. Conjeti, F. Tombari, N. Navab
Leveraging Web Data for Skin Lesion Classification Proceedings of the Bildverarbeitung für die Medizin 2019 (bib) |
|
| 2018 | |
| M. Paschali, S. Conjeti, F. Navarro, N. Navab
Generalizability vs. Robustness of medical imaging networks 21st International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Spain 2018. The original publication is available online at link.springer.com (bib) |
|
| F. Navarro, S. Conjeti, F. Tombari, N. Navab
Webly Supervised Learning for Skin Lesion Classification Proceedings of the In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2018, Granada, Spain. (bib) |
|
| S. Conjeti, M. Paschali, A. Guha Roy, N. Navab
Deep Hashing for Large-Scale Medical Image Retrieval Bildverarbeitung für die Medizin 2018 - Algorithmen - Systeme - Anwendungen. Proceedings des Workshops vom 11. bis 13. März 2018 in Erlangen The original publication is available online at link.springer.com (bib) |
|
| 2017 | |
| A. Guha Roy, S. Conjeti, D. Sheet, A. Katouzian, N. Navab, C. Wachinger
Error Corrective Boosting for Learning Fully Convolutional Networks with Limited Data Proceedings of MICCAI 2017: Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017 (bib) |
|
| S. Conjeti, M. Paschali, A. Katouzian, N. Navab
Deep Multiple Instance Hashing for Scalable Medical Image Retrieval The first two authors contributed equally. 20th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Canada 2017. The original publication is available online at link.springer.com (bib) |
|
| A. Kazi, S. Conjeti, A. Katouzian, N. Navab
Coupled Manifold Learning for Retrieval Across Modalities In Computer Vision Workshop (ICCVW), 2017 IEEE International Conference on (pp. 1321-1328). IEEE (bib) |
|
| 2016 | |
| S. Pölsterl, L. Wang, , S. Conjeti, A. Katouzian, N. Navab
Heterogeneous ensembles for predicting survival of metastatic, castrate-resistant prostate cancer patients F1000Research vol. 5, no. 2676, 2016 (bib) |
|
| S. Conjeti, A. Katouzian, A. Kazi, S. Mesbah, D. Beymer, T.F. Syeda Mahmood, N. Navab
Metric Hashing Forests Medical Image Analysis, Special Issue MICCAI 2015, Best Paper Award, 2016. (bib) |
|
| S. Pölsterl, S. Conjeti, N. Navab, A. Katouzian
Survival analysis for high-dimensional, heterogeneous medical data: Exploring feature extraction as an alternative to feature selection Artificial Intelligence in Medicine, vol. 72, pp. 1-11, 2016 (bib) |
|
| A. Shah, S. Conjeti, N. Navab, A. Katouzian
Deeply learnt hashing forests for content based image retrieval in prostate MR images SPIE Medical Imaging, San Diego, California, USA, March 2016 (bib) |
|
| S. Conjeti, S. Mesbah, M. Negahdar, P. Rautenberg, , N. Navab, A. Katouzian
Neuron-Miner: An Advanced Tool for Morphological Search and Retrieval in Neuroscientific Image Databases Neuroinformatics (Accepted March 2016). (bib) |
|
| S. Conjeti, A. Katouzian, A. Guha Roy, L. Peter, D. Sheet, S. Carlier, A. Laine, N. Navab
Supervised Domain Adaptation of Decision Forests: Transfer of models trained in vitro for in vivo intravascular ultrasound tissue characterization Medical Image Analysis, 2016 (bib) |
|
| 2015 | |
| S. Conjeti, S. Mesbah, A. Kumaraswamy, P. Rautenberg, N. Navab, A. Katouzian
Hashing forests for morphological search and retrieval in neuroscientific image databases Proceedings of the 18th International Conference on Medical Image Computing and Computer Assisted Interventions (MICCAI), Munich, Germany, October 2015 (bib) |
|
| S. Conjeti, S. Mesbah, A. Kumaraswamy, P. Rautenberg, N. Navab, A. Katouzian
Hashing forests for morphological search and retrieval in neuroscientific image databases Proceedings of the 18th International Conference on Medical Image Computing and Computer Assisted Interventions (MICCAI), Munich, Germany, October 2015 (bib) |
|
| S. Albarqouni, M. Baust, S. Conjeti, A. Al-Amoudi, N. Navab
Multi-scale Graph-based Guided Filter for De-noising Cryo-Electron Tomographic Data Proceedings of the British Machine Vision Conference (BMVC), pages 17.1-17.10. BMVA Press, September 2015 (bib) |
|
| A. Guha Roy, S. Conjeti, S. Carlier, P. K. Dutta, A. Kastrati, A. Laine, N. Navab, A. Katouzian, D. Sheet
Lumen Segmentation in Intravascular Optical Coherence Tomography using Backscattering Tracked and Initialized Random Walks IEEE Journal of Biomedical and Health Informatics, 2015 (In Press) (bib) |
|
| A. Guha Roy, S. Conjeti, S. Carlier, A. Konig, A. Kastrati, P. K. Dutta, A. Laine, N. Navab, D. Sheet, A. Katouzian
Bag of forests for modelling of tissue energy interaction in optical coherence tomography for atherosclerotic plaque susceptibility assessment Proceedings of International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, April 2015 (bib) |
|
| S. Conjeti, M. Yigitsoy, D. Sheet, J. Chatterjee, N. Navab, A. Katouzian
Mutually Coherent Structural Representation for Image Registration through Joint Manifold Embedding and Alignment Proceedings of International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, April 2015 (bib) |
|
| S. Conjeti, M. Yigitsoy, T. Peng, D. Sheet, J. Chatterjee, C. Bayer, N. Navab, A. Katouzian
Deformable Registration of immunofluorescence and Histology using iterative Cross-modal Propagation Proceedings of International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, April 2015 (bib) |
|
| 2014 | |
| T. Peng, L. Wang, C. Bayer, S. Conjeti, M. Baust, N. Navab
Shading Correction for Whole Slide Image Using Low Rank and Sparse Decomposition International Conference on Medical Image Computing and Computer Assisted Interventions (MICCAI), Boston, USA, September 2014 (bib) |
|
| , S. Conjeti, P. B. Noël, S. Carlier, N. Navab, A. Katouzian
Full-Wave Intravascular Ultrasound Simulation from Histology Medical Image Computing and Computer-Assisted Intervention, MICCAI, 2014 (bib) |
|
| 2013 | |
| D. Sheet, S. P. K. Karri, S. Conjeti, S. Ghosh, J. Chatterjee, A. K. Ray
Detection of retinal vessels in fundus images through transfer learning of tissue specific photon interaction statistical physics Proceedings of International Symposium on Biomedical Imaging (ISBI), San Francisco, CA, USA, April 2013 (bib) |
|
| 2011 | |
| R. R. Singh, S. Conjeti, R. Banerjee
An approach for real-time stress-trend detection using physiological signals in wearable computing systems for automotive drivers Intelligent Transportation Systems (ITSC), 2011 14th International IEEE Conference on (bib) |
|
Teaching Assistance
- Master Praktikum: Machine Learning in Medical Imaging WiSe? 2016/17
- Master Praktikum: Machine Learning in Medical Imaging SoSe? 2016
- Master Praktikum: Machine Learning in Medical Imaging WiSe? 2015/16
- Master Hauptseminar: Machine Learning in Imaging and Visualization, WiSe? 2014/15
- Lectures: Computer Aided Medical Procedures II, SoSe? 2015
| UsersForm | |
|---|---|
| Title: | Dr. |
| Circumference of your head (in cm): | |
| Firstname: | Sailesh |
| Middlename: | |
| Lastname: | Conjeti |
| Picture: | https://scholar.google.de/citations?view_op=medium_photo&user=C4eOUHsAAAAJ&citpid=5 |
| Birthday: | 15 December 1990 |
| Nationality: | India |
| Languages: | English |
| Groups: | Registration/Visualization, Segmentation, Medical Imaging, Machine Learning for Medical Applications, Microscopic Image Analysis |
| Expertise: | Registration/Visualization, Medical Imaging, Molecular Imaging |
| Position: | Scientific Staff |
| Status: | Alumni |
| Emailbefore: | sailesh.conjeti |
| Emailafter: | tum.de |
| Room: | MI 03.13.056 |
| Telephone: | +49 89 289 19400 |
| Alumniactivity: | |
| Defensedate: | |
| Thesistitle: | |
| Alumnihomepage: | |
| Personalvideo01: | |
| Personalvideotext01: | |
| Personalvideopreview01: | |
| Personalvideo02: | |
| Personalvideotext02: | |
| Personalvideopreview02: | |