Sep OsiriX Segmentation plugin
student: Brian JensenAdvisor: Prof. Dr. Nassir Navab
Supervision by: Ralph Bundschuh
Abstract
 OsiriX is an open source DICOM image viewer for Mac OS X offering advanced image visualization capabilities. The goal of this project was to create a plugin for the OsiriX DICOM viewer in order to add support for semi automatic segmentation of tumors in fused PET / CT images. After accepting the selection from a user the plugin offers the ability to perform several different kinds of region growing algorithms. The plugin can perform the segmentation on the image itself, or optionally on a registered image, such as a PET image in CT/ PET fused image setup, after which the segmented volume is then mapped back onto the main image. The plugin also provides methods simple integration with other image processing frameworks.
This project was conducted as a cooperation between the Chair for Computer Aided Medical Procedures and Augmented Reality at the TU München, and Department of Nuclear Medicine at the Univserity Hospital Rechts der Isar, TU München.
OsiriX is an open source DICOM image viewer for Mac OS X offering advanced image visualization capabilities. The goal of this project was to create a plugin for the OsiriX DICOM viewer in order to add support for semi automatic segmentation of tumors in fused PET / CT images. After accepting the selection from a user the plugin offers the ability to perform several different kinds of region growing algorithms. The plugin can perform the segmentation on the image itself, or optionally on a registered image, such as a PET image in CT/ PET fused image setup, after which the segmented volume is then mapped back onto the main image. The plugin also provides methods simple integration with other image processing frameworks.
This project was conducted as a cooperation between the Chair for Computer Aided Medical Procedures and Augmented Reality at the TU München, and Department of Nuclear Medicine at the Univserity Hospital Rechts der Isar, TU München.
Methods

 OsiriX is a native Mac OS X application and as such is mainly written in objective-c. The main image processing framework used in the plugin is the Insight Segmentation and Registration Toolkit (ITK) which is written using C++. Because of these constraints most of the plugin was written in objective-c++ (the interface and OsiriX interaction), and the segmentation code was written in pure C++. In addition a shell script was written for the automatic refactoring of the ITK namespace.
The user interface has support for three standard ITK region growing algorithms: connected threshold, neighborhood connected threshold and confidence connected. A fourth algorithm was developed using the ITK framework, gradient threshold. This custom threshold algorithm operates on image gradient magnitudes in the image, segmenting voxels whose gradient magnitude falls below a certain ratio starting with from a seed point.
OsiriX is a native Mac OS X application and as such is mainly written in objective-c. The main image processing framework used in the plugin is the Insight Segmentation and Registration Toolkit (ITK) which is written using C++. Because of these constraints most of the plugin was written in objective-c++ (the interface and OsiriX interaction), and the segmentation code was written in pure C++. In addition a shell script was written for the automatic refactoring of the ITK namespace.
The user interface has support for three standard ITK region growing algorithms: connected threshold, neighborhood connected threshold and confidence connected. A fourth algorithm was developed using the ITK framework, gradient threshold. This custom threshold algorithm operates on image gradient magnitudes in the image, segmenting voxels whose gradient magnitude falls below a certain ratio starting with from a seed point.

|
Plugin Documentation: NMSegmentation Documentation.pdf
Project Configuration Guide: ProjectConfiguration.pdf |
|---|
|
|
Final Presentation: Presentation.pdf |
|---|
Download
Latest release: 14.07.09

|
NMSegmention 1.01 binary (x86_64, i386, ppc) |
|---|
|
|
NMSegmentation 1.0.1 Source code |
|---|
Results
Here are some screenshots of the OsiriX plugin NMSegmentation in action.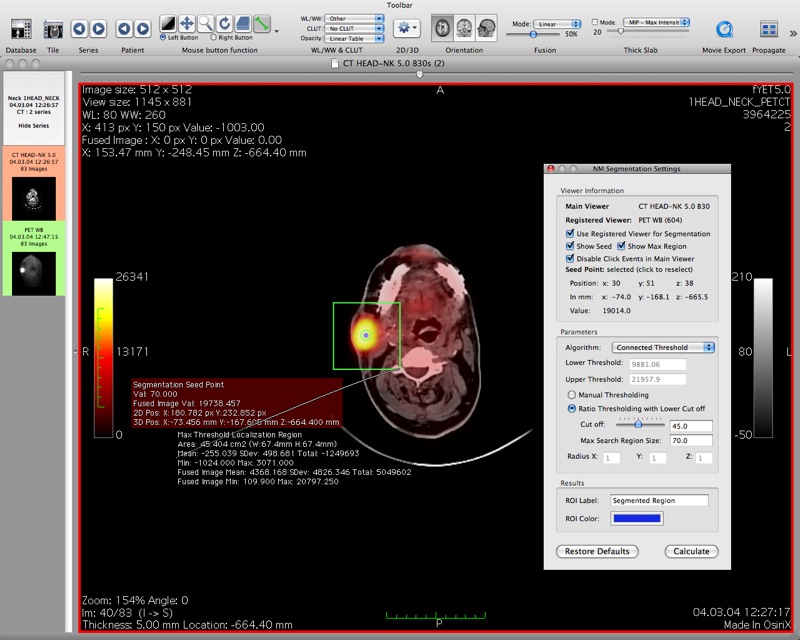
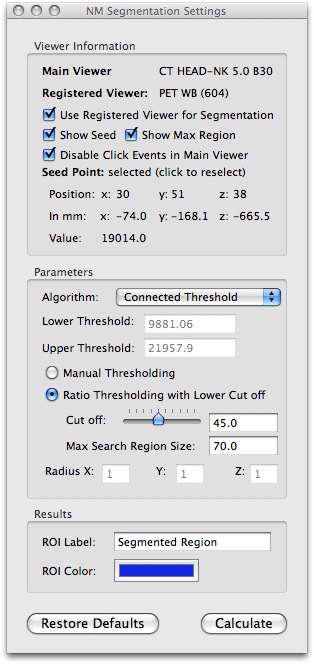
This screenshot shows the results after performing a connected threshold segmentation on the region of interest
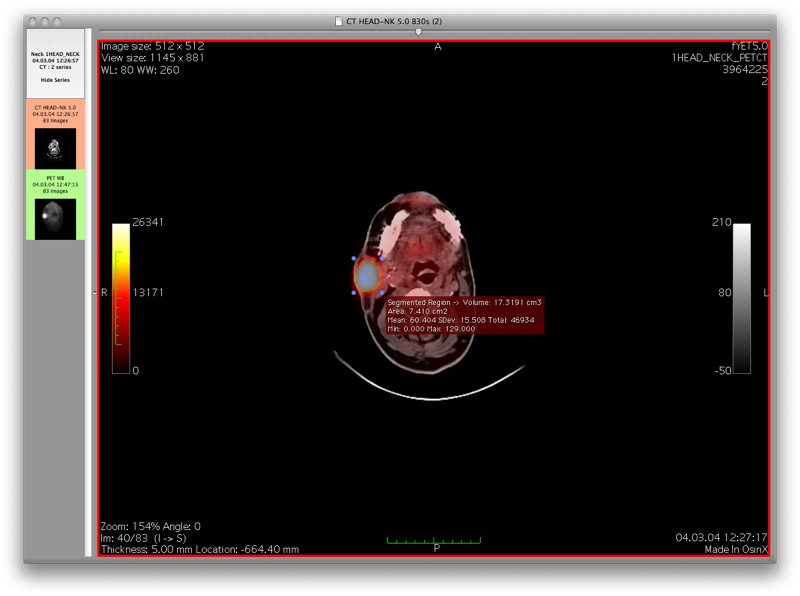
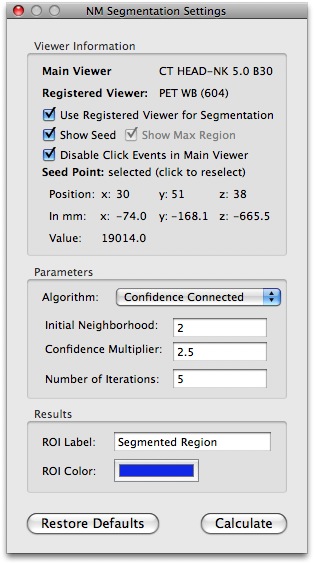
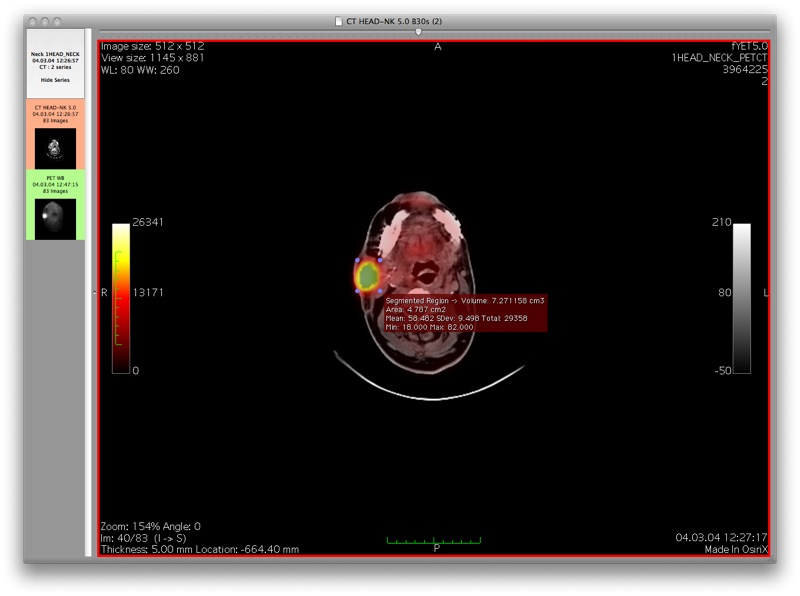
This screenshot shows the results using the gradient magnitude threshold segmentation algorithm on the same region
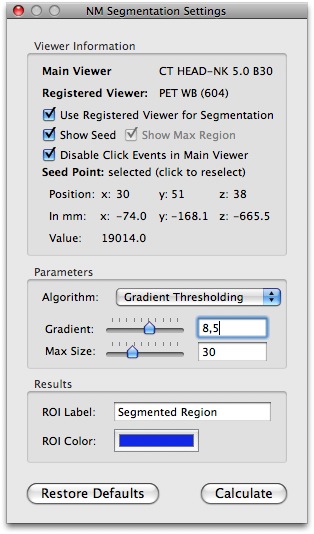
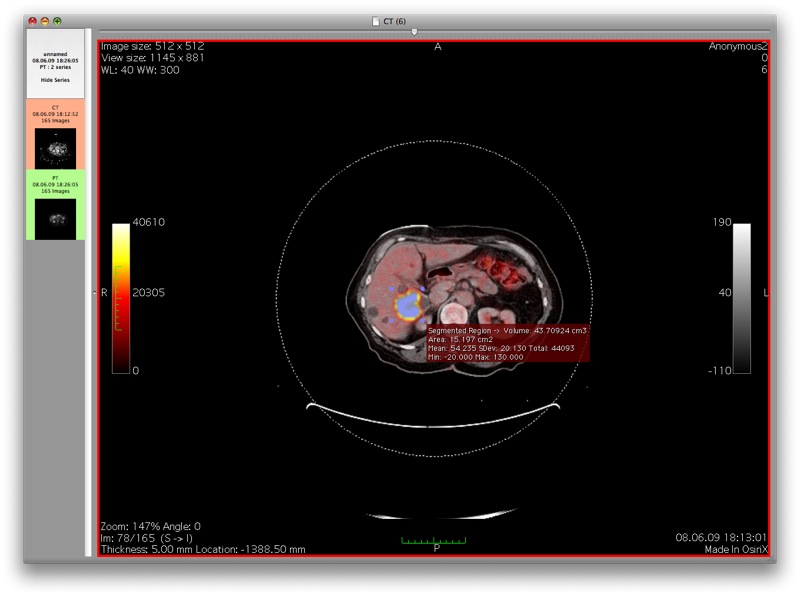
 This screenshot shows the results of the gradient threshold segmentation algorithm on a different lesion type
This screenshot shows the results of the gradient threshold segmentation algorithm on a different lesion type
| Students.ProjectForm | |
|---|---|
| Title: | Development of an OsiriX Segmentation plugin |
| Abstract: | OsiriX is an open source DICOM image viewer for Mac OS X offering advanced image visualization capabilities. The goal of this project is to create a plugin for the segmentation of tumors in fused PET / CT image. The plugin should provide methods for integrating with external image procesing libraries, so that additional segmentation can be easily added. |
| Student: | Brian Jensen |
| Director: | Nassir Navab |
| Supervisor: | Ralph Bundschuh |
| Type: | SEP |
| Area: | Segmentation, Medical Imaging, Molecular Imaging |
| Status: | finished |
| Start: | 05/2008 |
| Finish: | 06/2009 |