Histology Fold Detection
This project is part of a larger project that aims at the registration of histology volumes to in-vivo volumes. Before the registration is performed, first a histology volume needs to be reconstructed from histological slices. The reconstruction of histology volumes is performed in three stages- embedding of a sample in paraffin block
- slicing (cutting) the block and imaging the slices
- reconstructing a 3D volume from the 2D slices
Goals
One of the difficulties encountered in reconstruction is that the slicing causes deformations, shears and tears and folds in the slices, which introduce "inconsistencies" between subsequent slices. The goal of this clinical project, idp or bachelor/master thesis is to detect the folds and tears in the corrupted images. Examples of a good and a corrupted slice are shown in the figure: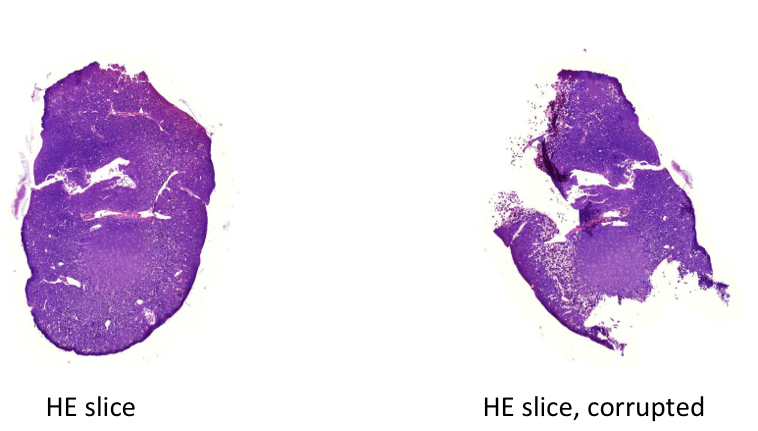
Requirements
- knowldedge in matlab and or C++
- previous knowledge in machine learning is a plus
Tasks:
- Develop a classification method that given a new slice it determines whether it is corrupted or not
- Develop an algorithm to detect the folds and tears in a corrupted image
Contact
- Diana Mateus : mateus@in.tum.de
- Martin Groher: groher@in.tum.de
| ProjectForm | |
|---|---|
| Title: | Histology Fold Detection |
| Abstract: | This project is part of a larger project that aims at the registration of histology volumes to in-vivo volumes. Before the registration is performed, first a histology volume needs to be reconstructed from histological slices. The reconstruction of histology volumes is performed in three stages first the embedding of a sample in paraffin block; second, the slicing (cutting) the block and imaging the slices; and third, the reconstructing a 3D volume from the 2D slices. One of the difficulties encountered in reconstruction is that the slicing causes deformations shears and tears and folds in the slices, which introduce "inconsistencies" between subsequent slices. The goal of this project is to classify the slices corrupted by such deformations and to detect the tears and folds in the image. |
| Student: | |
| Director: | Prof. Nassir Navab |
| Supervisor: | Martin Groher and Diana Mateus |
| Type: | DA/MA/BA |
| Area: | Registration / Visualization |
| Status: | draft |
| Start: | |
| Finish: | |
| Thesis (optional): | |
| Picture: | |
| I | Attachment  | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|
| | corrupted-slice.png | manage | 288.2 K | 03 May 2011 - 16:21 | DianaMateus | |
| | rat-kidney-reconstruction.png | manage | 144.7 K | 03 May 2011 - 16:21 | DianaMateus |