Multi-Organ Segmentation without registrationScientific Director: Vasileios Zografos, Bjoern MenzeContact Person(s): Vasileios Zografos |
Abstract
This project is concerned with the automatic segmentation of multiple organs in 3D medical images. Initially, we have investigated abdominal CT images but our scope will grow to full-body scans, multiple modalities and other structures (such as bones, muscles etc). Because we are not using expensive registration methods our approach is well suited to very large clinical studies with thousands of medical volumes.Detailed Project Description
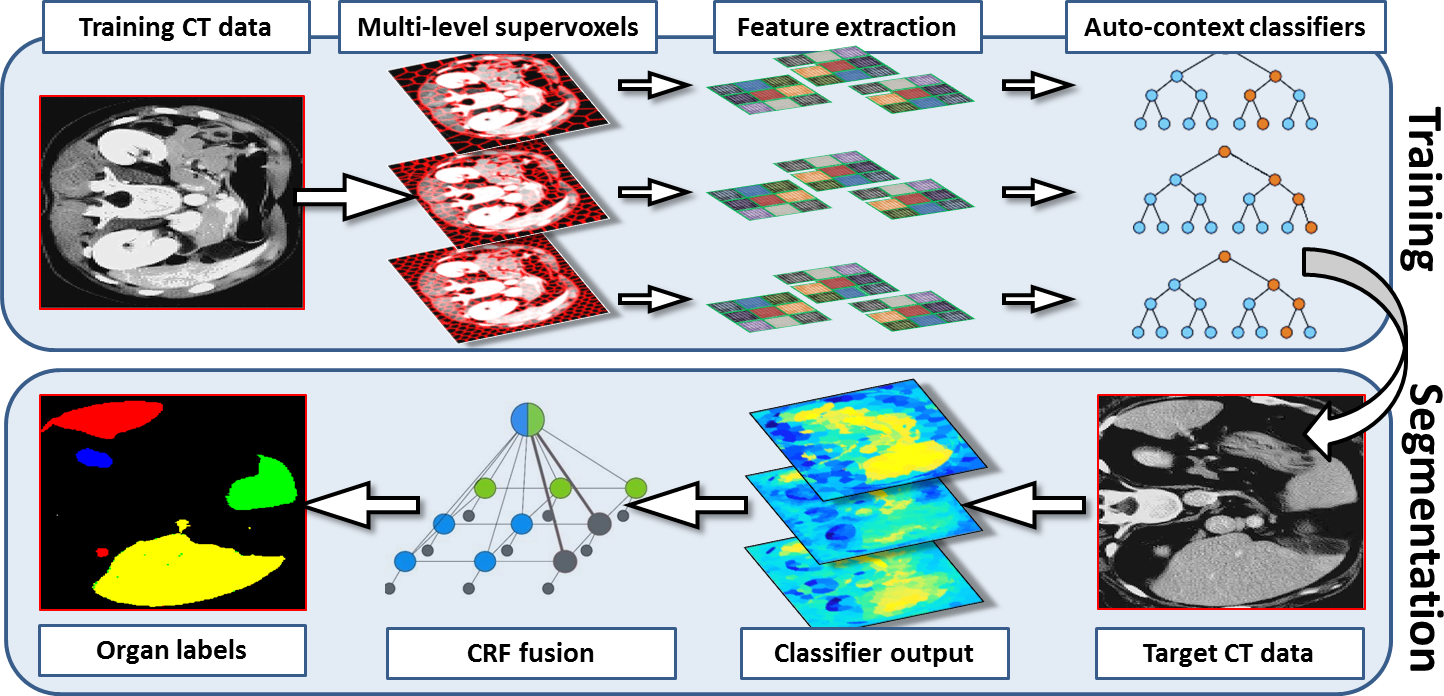
Most existing work in multi-organ segmentation is based on registration approaches. However, such approaches are slow , inaccurate when faced with large inter-subject variabilities and require all the data to be available during segmentation time.We propose a framework for multi-organ segmentation which leverages several ideas from computer vision and machine learning and does not require any registration during training or during segmentation time.
-- We first generate supervoxels from each CT image at multiple levels of detail.
-- We the extract a set of complimentary appearance and context features from the supervoxels.
-- We then train a discriminative model at each level (Gradient Boosted Tree)
-- The disriminative models are linked together in a hierarchical auto-context fashion.
-- Finally, the output is fused using a hierarchical CRF
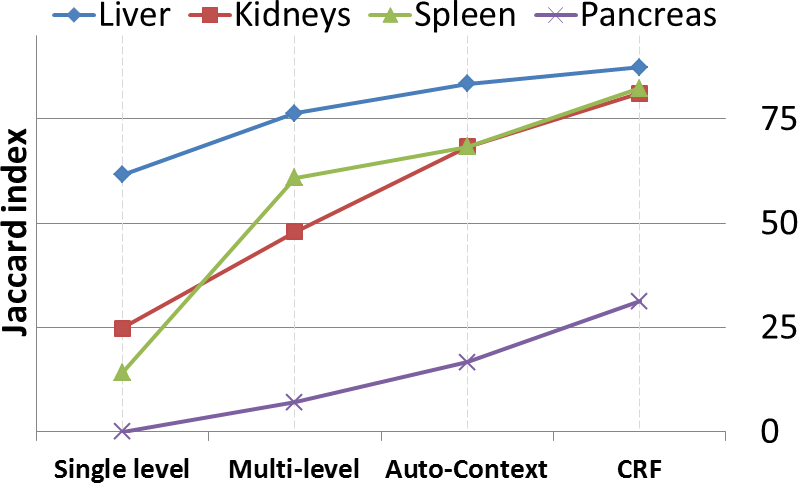
Our method is fast , accurate and because training is done offline and decoupled from the segmentation stage, we can increase accuracy by training with more data but wihout incurring any additional segmentation cost. In addition, we do not require the trained data (atlases) to be available during segmentation time. All we need to store is a small set of trained classifiers with a minimal memory footprint and without data storage and privacy issues. This makes our method efficient , portable and very practical.
Pictures
Team
Contact Person(s)
Location
| Technische Universität München Institut für Informatik / I16 Boltzmannstr. 3 85748 Garching bei München Tel.: +49 89 289-17058 Fax: +49 89 289-17059 |
internal project page
Please contact Vasileios Zografos for available student projects within this research project.