Assessment of molecular dynamics in brain tissue through accelerated diffusion and exchange imagingIn medical collaboration with:Max Plank Institute of Psychiatry In academic collaboration with: CUBRIC Scientific Director: Prof. Dr. Bjoern Menze Contact Person(s): Miguel Molina Romero In industrial collaboration with: General Electric Global Research |
Abstract
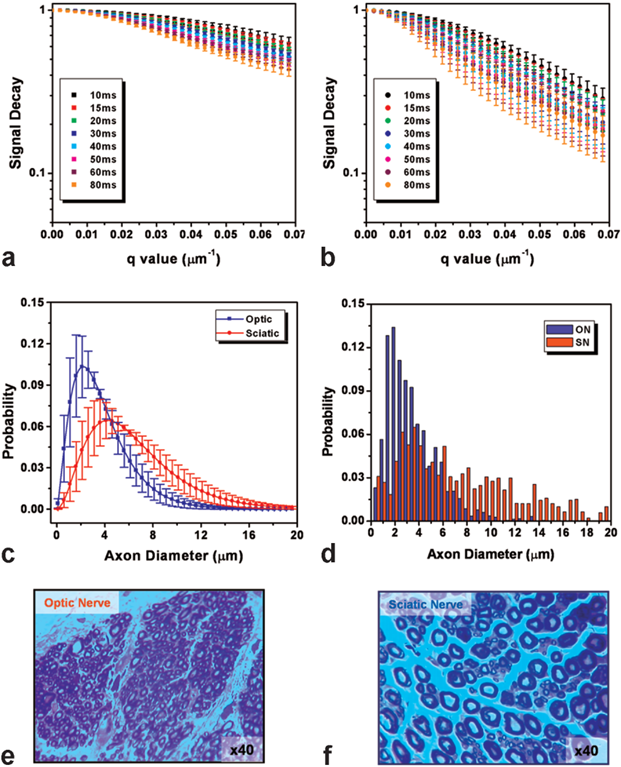
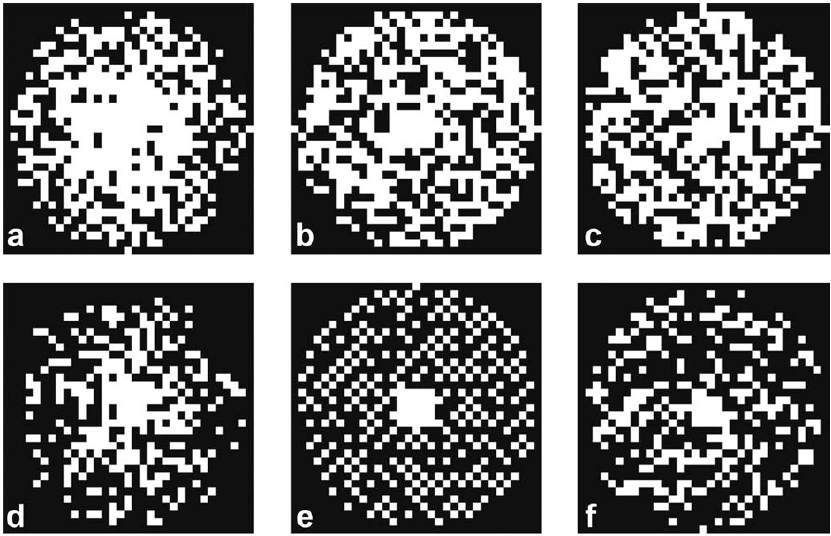
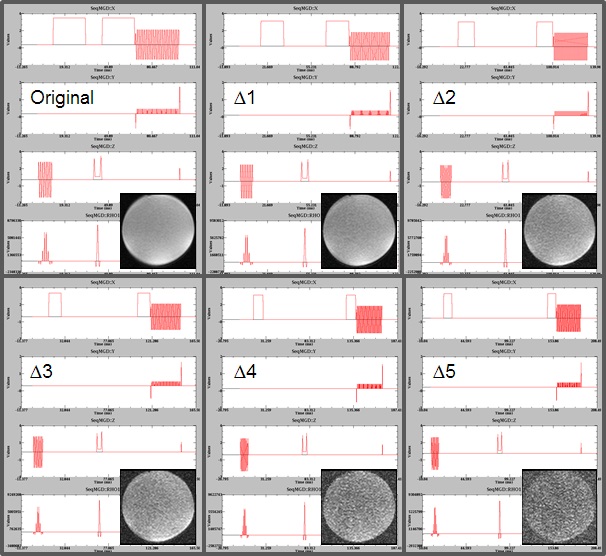
Diffusion of water molecules and spin-spin relaxation can be measured, in vivo, through MRI. These measurements contain information of the underlying tissue microstructure and thus, they are of interest. In this project we want to, first, assess these two contrasts within a clinical feasible time, using a modified pulse sequence and an accelerated acquisition scheme based on compressed sensing. Second, develop a model, or adapt an existing one, that allows one to extract tissue microstructural information out of the measurements. And third, validate the meaning of this information for brain diseases and brain plasticity in clinical and research environments.Pictures
|
|
Team
Contact Person(s)
|
Working Group
|
|
|
|
Location
| Technische Universität München Institut für Informatik / I16 Boltzmannstr. 3 85748 Garching bei München Tel.: +49 89 289-17058 Fax: +49 89 289-17059 |
internal project page
Please contact Miguel Molina Romero for available student projects within this research project.