Color Normalization for Histology Image Processing
Thesis by:Advisor: Prof. Dr. Nassir Navab
Supervision by: Dr. Tingying Peng, Dr. Lichao Wang
Project Description
Automated image processing and quantification are increasingly gaining attention in the field of digital pathology. However, a common problem in histology images is the color variation introduced by the use of different microscopies, scanners or inconsistencies in tissue preparation (an example of color variation is shown in Fig.1.). This creates difficulty in image interpretation by both pathologists and computational pathology software trained on a particular stain appearance.e.g. learning-based method. This project is to address the challenging issue of color inconsistency in histology and to develop a fast, accurate and robust color normalization technique. The difficulty of the project will be adapted to an IDP, Bachelor or Master Thesis.
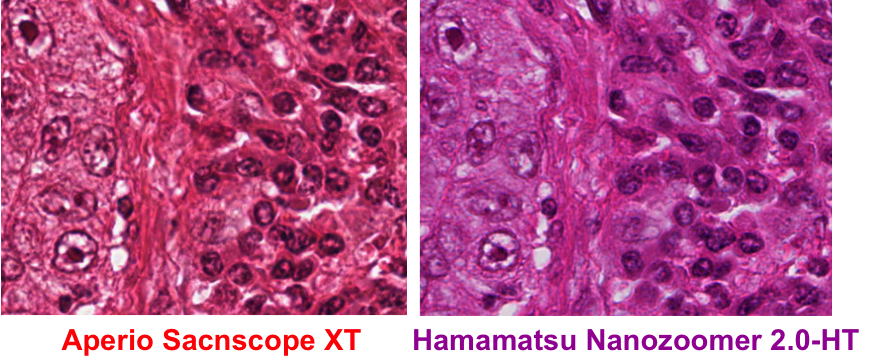 Figure 1: Same tissue section under different scanner (left) Aperio scanner (right) Hamamatsu scanner.
Figure 1: Same tissue section under different scanner (left) Aperio scanner (right) Hamamatsu scanner.
Tasks
Requirement
Literature[1] A. Vahadane, T. Peng, S. Albarqouni, M. Baust, K. Steiger, M. Schlitter, A. Sethi, N. Navab: "Structure-preserved Color Normalization For Histological Images". ISBI(2015).
| ||||||||||||||||||||||||||