Tingying Peng, Ph.D
 |
|
Education
- Ph.D. in Biomedical Engineering, University of Oxford, Oxford, UK
- B.Sc. in Electrical Engineering, Minor in Applied Mathematics, Peking University, Beijing, China
- Ph.D Thesis: "Signal processing methods for the analysis of cerebral blood flow and metabolism"
download
Awards
- Humboldt Postdoc Fellowship, Jan. 2013-Dec.2014.
- Dorothy Hodgkins Postgraduate Award, Oct. 2005-Sep. 2008.
- Junzheng Scientific Program Scholarship, Mar. 2003-Jun. 2004.
Research Interests
- Molecular imaging
- Microscopic image processing
- Machine learning for medical applications
- Mathematical modeling for physiological understanding
Active research projects
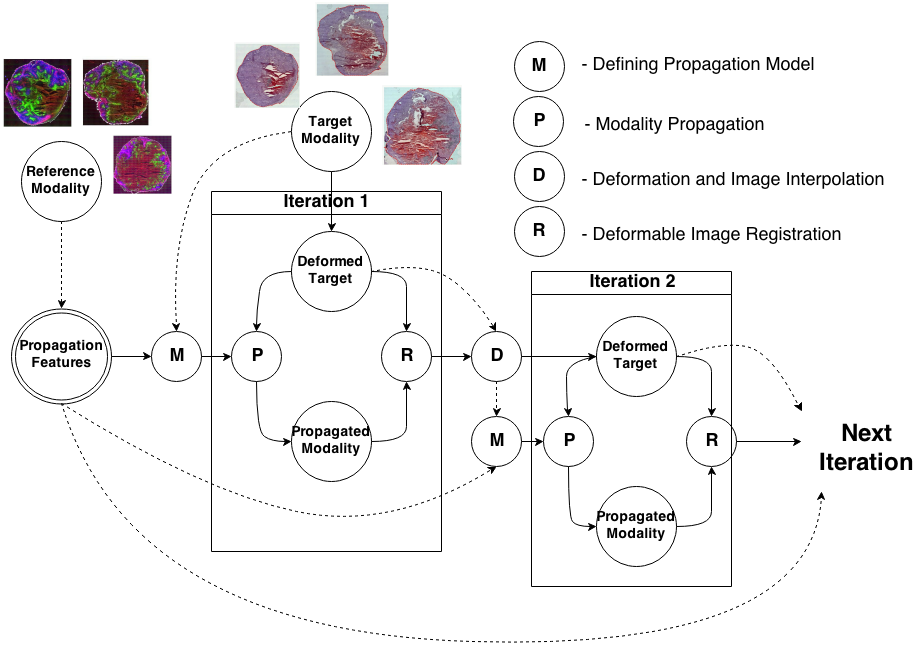
Knowledge Propagation Models for Image RegistrationTo register modalities with complex intensity relationships, we leverage machine learning algorithm to cast it into a mono modal registration problem. This is done by extracting tissue specific features for propagating anatomical/structural knowledge from one modalitiy to an other through an online learnt propagation model. The registration and propagation steps are iteratively performed and refined. For proof-of-concept, we employ it for registering (1) Immunofluorescence to Histology images and (2) Intravascular Ultrasound to Histology Images. |
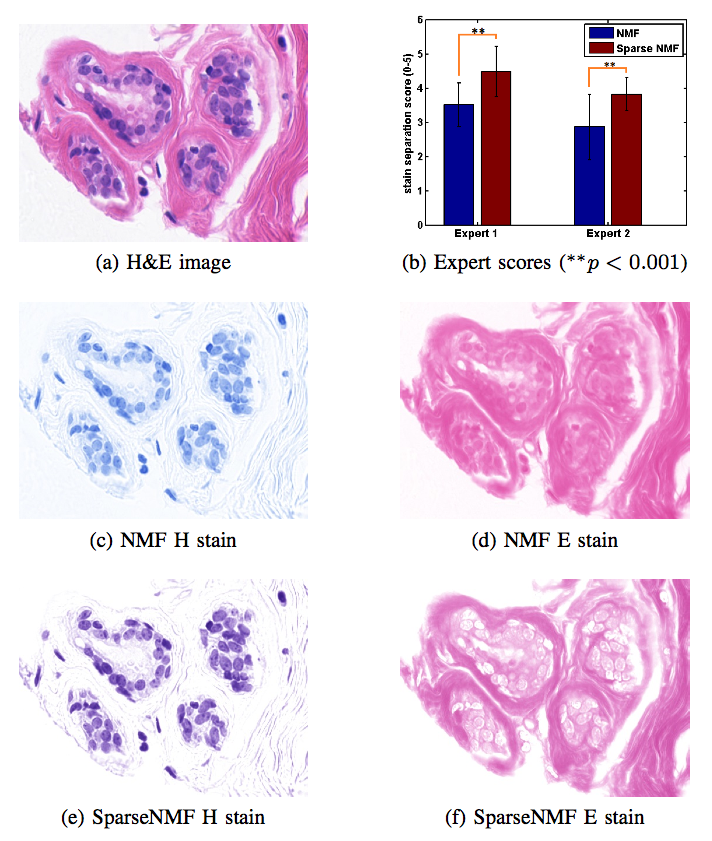
Stain Separation and Structure-Preserving Color Normalization for Histological ImagesStaining and scanning of tissue samples for microscopic examination is fraught with unwanted variations that affect their color appearance. Sources of these variations include differences in raw material and manufacturing techniques of stain vendors, staining protocols of labs, and color responses of digital scanners. Color normalization of stained biopsies and tissue microarrays will help pathologists and computational pathology software while comparing different tissue samples. However, techniques that are used for natural images, such as histogram matching fail to utilize unique properties of stained tissue samples and produce undesirable artifacts. Tissue samples are stained with only a few reagents (frequently only two -- hemotoxylin and eosin or H\&E) and most tissue regions bind to only one stain or the other, thus producing sparse density maps composed of only a few components. This underlying structure of sparse stain density is biomedically important. We used these properties of stained tissue to propose a technique for stain separation and color normalization. Based on sparse non-negative matrix factorization (sparseNMF), we estimate prototype color and density map of each stain in an unsupervised manner to perform stain separation. To color normalize a given source image, we combine its stain density maps with the stain color prototypes of a target image whose appearance was preferred by pathologists. In this way, the normalized image preserve the biological structure encoded in the stain density of the source image. Both the proposed sparseNMF stain separation and color-normalization techniques yield higher correlation with ground truth than the state of the art. They are also rated qualitatively higher than other techniques by a group of pathologists. We further propose a computationally faster extension of this technique for large whole-slide images that selects an appropriately small sample of patches to compute the color prototypes of each stain instead of using the entire image. The fast scheme achieves a 20-folds acceleration, which does not only greatly enhance the analysis efficiency, but also allow its clinical applications to become practically feasible. |
Open student projects
Please contact me.Publications
| 2020 | |
| A. Sadafi, A. Makhro, A. Bogdanova, N. Navab, T. Peng, S. Albarqouni, C. Marr
Attention based Multiple Instance Learning for Classification of Blood Cell Disorders International Conference on Medical Image Computing and Computer Assisted Interventions (MICCAI), Lima, Peru, 2020 (bib) |
|
| T. Peng, L. Lamm, M. Loeffler, N. Ahmed, N. Navab, T. Schroeder, C. Marr
Background and illumination correction for time-lapse microscopy data with correlated foreground International Conference on Medical Image Computing and Computer Assisted Interventions (MICCAI), Lima, Peru, 2020 (bib) |
|
| 2017 | |
| A. Keskin, M. Kekus, H. Adelsberger, U. Neumann, D. Shimshek, B. Song, B. Zott, T. Peng, H. Foerstl, M. Staufenbiel, I. Nelken, B. Sakmann, A. Konnerth, M. Busche
BACE inhibition-dependent repair of Alzheimer Ìs pathophysiology PNAS, in press (bib) |
|
| T. Peng, K. Thorn, T. Schroeder, L. Wang, F. Theis, C. Marr, N. Navab
A BaSiC? Tool for Background and Shading Correction of Optical Microscopy Images Nature Communications 8:14836 (bib) |
|
| G. Carneiro, T. Peng, C. Bayer, N. Navab
Automatic Quantification of Tumour Hypoxia from Multi-modal Microscopy Images using Weakly-Supervised Learning Methods IEEE Transactions on Medical Imaging PP(99):1-1 (bib) |
|
| 2016 | |
| G. Bortsova, M. Sterr, L. Wang, F. Milletari, N. Navab, A. Boettcher, H. Lickert, F. Theis, T. Peng
Mitosis Detection in Intestinal Crypt Images with Hough Forest and Conditional Random Fields 7th International Workshop on Machine Learning in Medical Imaging (MLMI), Athena, Greece, October 2016 (bib) |
|
| M. Mishra, S. Schmitt, L. Wang, M. Strasser, C. Marr, N. Navab, H. Zischka, T. Peng
Structure-based Assessment of Cancerous Mitochondria using Deep Networks International Symposium on Biomedical Imaging (ISBI): From Nano to Macro, Prague, Czech Republic, April 2016 (bib) |
|
| A. Vahadane, T. Peng, A. Sethi, S. Albarqouni, L. Wang, M. Baust, K. Steiger, A. M. Schlitter, I. Esposito, N. Navab
Structure-Preserving Color Normalization and Sparse Stain Separation for Histological Images IEEE Transactions on Medical Imaging (TMI), vol. 35, no. 8, pp. 1962 - 1971, 2016. (bib) |
|
| 2015 | |
| G. Carneiro, T. Peng, C. Bayer, N. Navab
Weakly-supervised Structured Output Learning with Flexible and Latent Graphs using High-order Loss Functions International Conference on Computer Vision (ICCV), Santiago, Chile, December 2015 (bib) |
|
| G. Carneiro, T. Peng, C. Bayer, N. Navab
Automatic Detection of Necrosis, Normoxia and Hypoxia in Tumors from Multimodal Cytological Images International Conference on Image Processing (ICIP), Quebec, Canada, September 2015 (bib) |
|
| G. Carneiro, T. Peng, C. Bayer, N. Navab
Flexible and Latent Structured Output Learning: Application to Histology. MICCAI Workshop Machine Learning in Medical Imaging (MLMI), Munich, Germany, September 2015 (bib) |
|
| A. Vahadane, T. Peng, S. Albarqouni, M. Baust, K. Steiger, A. M. Schlitter, A. Sethi, I. Esposito, N. Navab
Structure-Preserved Color Normalization for Histological Images International Symposium on Biomedical Imaging (ISBI): From Nano to Macro, New York, USA, April 2015 (bib) |
|
| S. Conjeti, M. Yigitsoy, T. Peng, D. Sheet, J. Chatterjee, C. Bayer, N. Navab, A. Katouzian
Deformable Registration of immunofluorescence and Histology using iterative Cross-modal Propagation Proceedings of International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, April 2015 (bib) |
|
| 2014 | |
| T. Peng, L. Wang, C. Bayer, S. Conjeti, M. Baust, N. Navab
Shading Correction for Whole Slide Image Using Low Rank and Sparse Decomposition International Conference on Medical Image Computing and Computer Assisted Interventions (MICCAI), Boston, USA, September 2014 (bib) |
|
| T. Peng, M. Yigitsoy, A. Eslami, C. Bayer, N. Navab
Deformable registration of multi-modal microscopic images using a pyramidal interactive registration- learning methodology 6th Workshop on Biomedical Image Registration, London, UK, July, 2014 (bib) |
|
Other Publications
| UsersForm | |
|---|---|
| Title: | Dr. |
| Circumference of your head (in cm): | |
| Firstname: | Tingying |
| Middlename: | |
| Lastname: | Peng |
| Picture: |  |
| Birthday: | 14.02.1984 |
| Nationality: | China |
| Languages: | English, German, Chinese |
| Groups: | |
| Expertise: | |
| Position: | Scientific Staff |
| Status: | Active |
| Emailbefore: | tingying.peng |
| Emailafter: | tum.de |
| Room: | MI 03.13.057 |
| Telephone: | +49 89 289 19400 |
| Alumniactivity: | |
| Defensedate: | |
| Thesistitle: | |
| Alumnihomepage: | |
| Personalvideo01: | |
| Personalvideotext01: | |
| Personalvideopreview01: | |
| Personalvideo02: | |
| Personalvideotext02: | |
| Personalvideopreview02: | |