Glioma Modelling Using Spectroscopy and PET Images
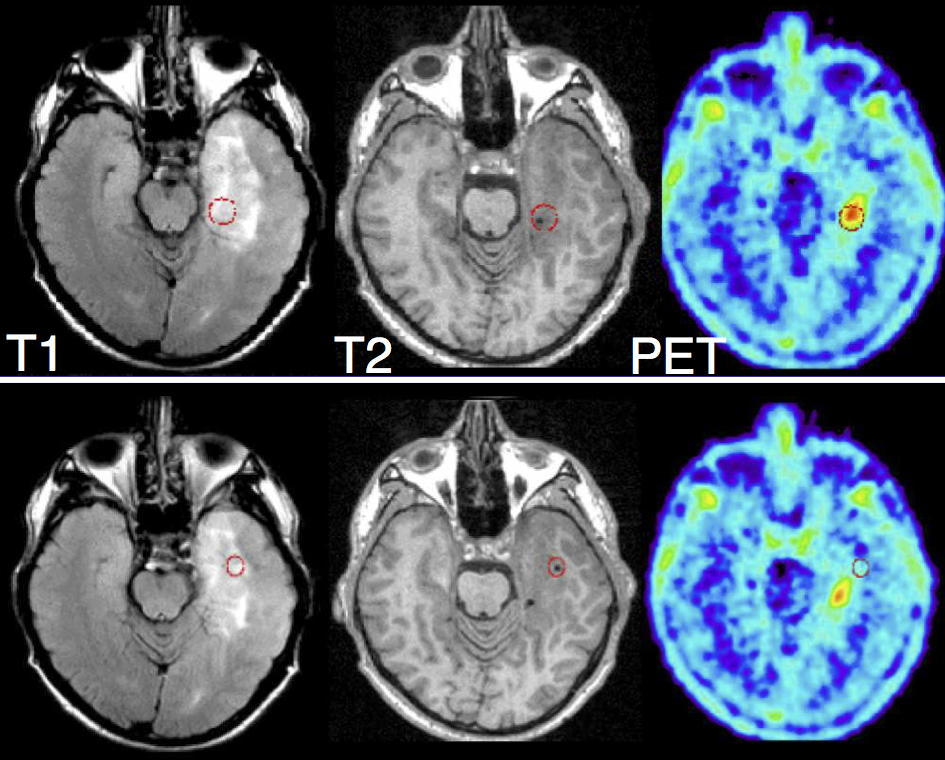 Figure 1: Red circles indicates two biopsy sites. The top case was proved positive for tumor, bottom
Figure 1: Red circles indicates two biopsy sites. The top case was proved positive for tumor, bottom
case negative. Only PET differentiated between the two sites. (Figure provided by Prof. Tony Lomax (PSI) and Frank Floeth (Uni Dusseldorf)
|
Project Description
Glioma is the most common type of primary brain tumors. It is characterised by infiltrative growth into the surrounding tissue, instead of forming a solid tumor with a well defined boundary. Due to this, only a part of the glioma beyond a certain concentration threshold is visible in MR images. In contrast, brain tissue infiltrated by tumor cells at lower concentrations appears normal on images obtained with current imaging modalities.
Several models were proposed to predict tumor distribution beyond threshold visible from MRI scans [1]. Most of these models describe tumor by a concentration field and simulate its evolution over time and space. For model personalization, patient specific model parameters need to be infer[2]. However parameter estimation based on mapping simulated tumor concentration with binary tumor segmentation might lead to a large set of possible parameters, all corresponding to the similar tumor structure but various concentration. Nonetheless amount of tissue infiltration is a leading factor in radiotherapy planing.
Even MRI provides superior structural information, imaging modalities like Magnetic Resonance Spectroscopy (MRS) and Positron Emission Tomography (PET) provides additional information (see Figure 1). Both PET and MRS measure metabolic activities in normal and tumor tissue, which are related to tumor concentration.
Tasks
- Incorporate information from PET and MRS in order to estimate tumor concentration
- Integration of the estimated concentration in existing glioma solver (C++)
- Validation of the model on clinical data
|
|
Requirements
- Good Programming Skills (C++)
- Basics of Numerical Analysis
- Desire to Learn and Improvise
- Independent worker
Contact
If you are interested in the project or if you have any questions please contact
Detail Project Description & References
- More information about the project together with corresponding references can be found on TUM internal page:
- If you have any problem with accessing the internal page, please let me (Jana Lipkova) know and I will be happy to help you


 Figure 1: Red circles indicates two biopsy sites. The top case was proved positive for tumor, bottom
Figure 1: Red circles indicates two biopsy sites. The top case was proved positive for tumor, bottom