Intraoperative 2D-3D Registration for Knee Alignment Surgery
Advisor: Alexander "Amazing Alex" Winkler, Matthias "Marvellous Matthias" GrimmSupervision by: Nassir Navab
Abstract
In this project a system is developed, which assists surgeons in verifying a surgical result by comparing interventional X-Rays to a preoperative plan. The targeted surgical procedure is knee osteotomy in which a bone (usually the tibia) is cut at a specific point and the two segments are repositioned to correct knee alignment. The 2D X-Ray images are to be registered to the 3D preoperative plan in order to compute the achieved 3D configuration between the two bone segments. This allows the surgeon to make sure that the plan is carried out accurately, or make adjustments if necessary, as correct knee alignment leads to improved patient outcome.[1]Introduction and Related Works
Knee osteotomy is a surgical procedure for managing different knee deformities. It corrects poor knee alignment and is frequently used when a patient has damaged only one side of the knee joint. By shifting weight off of the damaged side of the joint, pain can be relieved and significantly improve function in an arthritic knee. During this procedure, a cut is made on either the femur or tibia bone in the vicinity of the knee and either a wedge of bone is removed and then the bones are brought together (closing wedge osteotomy) or the wedge of bone is opened and a bone graft is added to fill the space (opening wedge osteotomy) as seen in Fig. 1.[2]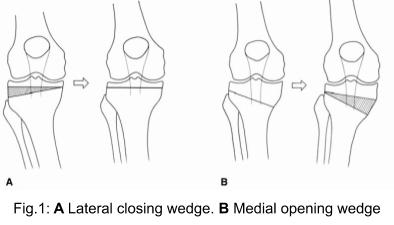 Traditionally the preoperative planning is performed on multiple radiographic views. Lower limb alignment can be assessed from full length radiographs of the lower extremities. Calculation of the correction angle is often done directly on the Anteroposterior weight-bearing radiographs. However deciding on a plan only in a frontal view can result in unwanted changes of the anatomy in the dimensions as well, as for example the posterior tibial slope.[3] Therefore planning can alternatively be done in 3D, such as based on Magnetic Resonance Imaging (MRI) or Computed Tomography (CT) data. During the procedure the surgeon tries to implement this plan and verifies the achieved alignment based on intraoperative X-Ray images. There is however a disconnect between the 2D X-Ray images and the 3D plan: in clinical practise the surgeon only inspects the 2D images and performs measurements directly on them, without automatic translation of the information from the 3D plan.
Registration between the intraoperative X-rays and the plan can be used to overcome these issues. A doctor could acquire an X-ray, and then an algorithm would compute the 6 degree of freedom pose between the C-arm (taking the X-Ray) and the preoperative plan. This enables a mapping between intraoperative X-ray and the pre-operative plan. Therefore registration allows the surgeon to properly assess the situation during the surgery and take appropriate measures, if necessary.
Traditionally the preoperative planning is performed on multiple radiographic views. Lower limb alignment can be assessed from full length radiographs of the lower extremities. Calculation of the correction angle is often done directly on the Anteroposterior weight-bearing radiographs. However deciding on a plan only in a frontal view can result in unwanted changes of the anatomy in the dimensions as well, as for example the posterior tibial slope.[3] Therefore planning can alternatively be done in 3D, such as based on Magnetic Resonance Imaging (MRI) or Computed Tomography (CT) data. During the procedure the surgeon tries to implement this plan and verifies the achieved alignment based on intraoperative X-Ray images. There is however a disconnect between the 2D X-Ray images and the 3D plan: in clinical practise the surgeon only inspects the 2D images and performs measurements directly on them, without automatic translation of the information from the 3D plan.
Registration between the intraoperative X-rays and the plan can be used to overcome these issues. A doctor could acquire an X-ray, and then an algorithm would compute the 6 degree of freedom pose between the C-arm (taking the X-Ray) and the preoperative plan. This enables a mapping between intraoperative X-ray and the pre-operative plan. Therefore registration allows the surgeon to properly assess the situation during the surgery and take appropriate measures, if necessary.
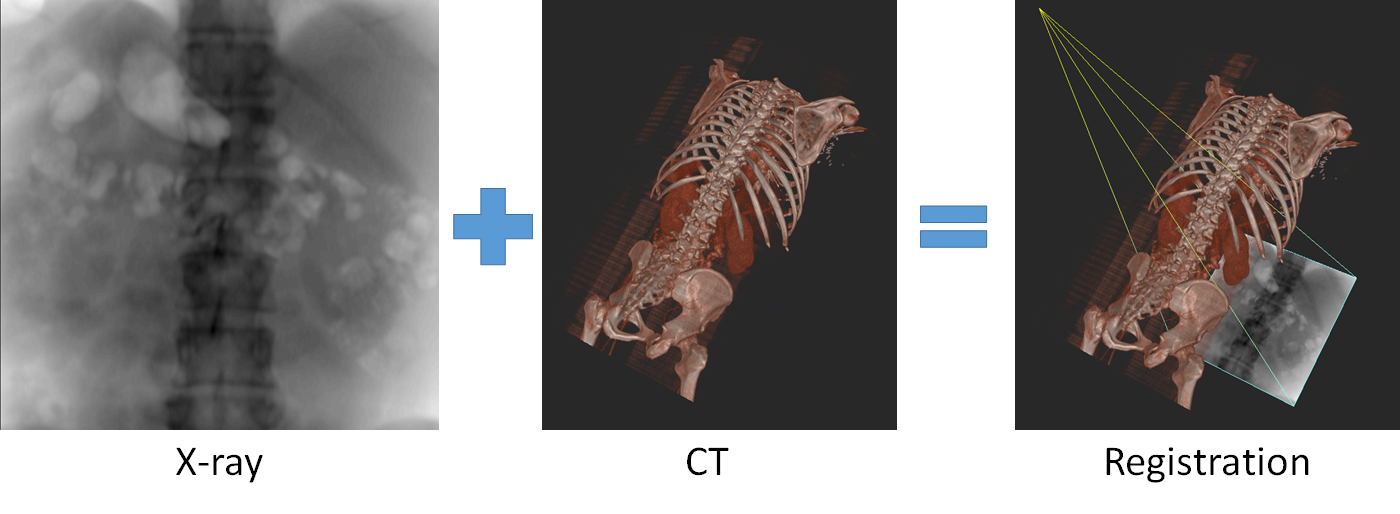 X-Ray to CT registration is a wide field of research. There are two basic families of methods. Feature-based methods rely on identifying corresponding anatomical landmarks in the X-Ray and in the CT. Then the corresponding perspective-n-point problem[4] can be solved to compute the corresponding pose. In practice, the correspondences are computed using markers. Marker-based methods suffer from problems though. Bone-implanted markers require a second, invasive procedure, whereas skin-implanted markers are susceptible to deformation.[5]
A second family of registration methods are intensity-based registration algorithms. These algorithms start from an initial pose. Then, the ideal registration is computed by iteratively optimizing a similarity metric between the real X-ray and a virtual X-ray (a digitally reconstructed radiograph; DRR).
The difference between these two methods is the following: Intensity-based methods are very precise, but they require an initialization. Feature point based methods do not require an initialization, but they are less precise.
Rather than relying on markers, landmarks can also be computed using Deep Learning. Hence it is a natural step to combine a feature based initialization with a intensity-based fine-tuning step.[6]
So far, all the discussed methods are rigid registration methods. These only compute the pose of the C-arm, assuming that there is no deformation between the time when the CT was acquired and the time when the X-ray was acquired. Some methods model the registration problem as registering several anatomical components individually in a rigid manner. Components could be Pelvis and femur for example. This allows to estimate the deformation between the movable limbs, while assuming the limbs themselves to be rigid.[7]
All of the aforementioned methods have one drawback, though. They assume that a network that is trained on DRRs can perform equally on real X-rays as it can perform on DRRs. In practice, this is not the case. A recent study shows that even when using state of the art DRR generators, the performance of a network decreases by a factor of four, when used on real X-rays, as opposed to when used on DRRs, assuming the network has only been trained on DRRs.[8] A recent method aims to overcome this issue. The method is inspired by the domain randomization works from robotics. Its idea is to enable a network trained on DRRs to run equally well on real X-rays, by increasing the variability of the DRRs during training as much as possible. This is done by both using multiple DRR generators, as well as using an elaborate randomized post-processing scheme.[9]
X-Ray to CT registration is a wide field of research. There are two basic families of methods. Feature-based methods rely on identifying corresponding anatomical landmarks in the X-Ray and in the CT. Then the corresponding perspective-n-point problem[4] can be solved to compute the corresponding pose. In practice, the correspondences are computed using markers. Marker-based methods suffer from problems though. Bone-implanted markers require a second, invasive procedure, whereas skin-implanted markers are susceptible to deformation.[5]
A second family of registration methods are intensity-based registration algorithms. These algorithms start from an initial pose. Then, the ideal registration is computed by iteratively optimizing a similarity metric between the real X-ray and a virtual X-ray (a digitally reconstructed radiograph; DRR).
The difference between these two methods is the following: Intensity-based methods are very precise, but they require an initialization. Feature point based methods do not require an initialization, but they are less precise.
Rather than relying on markers, landmarks can also be computed using Deep Learning. Hence it is a natural step to combine a feature based initialization with a intensity-based fine-tuning step.[6]
So far, all the discussed methods are rigid registration methods. These only compute the pose of the C-arm, assuming that there is no deformation between the time when the CT was acquired and the time when the X-ray was acquired. Some methods model the registration problem as registering several anatomical components individually in a rigid manner. Components could be Pelvis and femur for example. This allows to estimate the deformation between the movable limbs, while assuming the limbs themselves to be rigid.[7]
All of the aforementioned methods have one drawback, though. They assume that a network that is trained on DRRs can perform equally on real X-rays as it can perform on DRRs. In practice, this is not the case. A recent study shows that even when using state of the art DRR generators, the performance of a network decreases by a factor of four, when used on real X-rays, as opposed to when used on DRRs, assuming the network has only been trained on DRRs.[8] A recent method aims to overcome this issue. The method is inspired by the domain randomization works from robotics. Its idea is to enable a network trained on DRRs to run equally well on real X-rays, by increasing the variability of the DRRs during training as much as possible. This is done by both using multiple DRR generators, as well as using an elaborate randomized post-processing scheme.[9]
Task
Students should develop a method that takes as input one or multiple intraoperative X-rays and a preoperative plan (3d model) and use them to register the X-ray to separate parts of the plan. To be more precise, they should first register the X-ray to the upper part of the Tibia model and then register the X-ray to the lower part of the Tibia model. This enables to compute the relative transformation between the segments. Students will get access to preoperative plans and X-ray acquisitions from multiple cadavers using a tracked C-arm. The tracking system provides them with the relative pose of all the acquisitions to each other. In order to convert this to a ground truth, they would need to manually register one of the X-rays per cadaver and then use the tracked poses to compute the ground truth for each acquisition. The students will also receive access to the ImFusion? SDK, which can be used for this project, which already provides a multitude of functionality such as reading medical datasets and some registration functionality as a C++ and Python API. Based on the number of students working on the project and the pace of the progress, tasks for efficient intraoperative visualization or conducting a user study with surgeons may optionally be added to the work load.Requirements
Required:- Basic knowledge of image registration
- Basic knowledge of image processing
- Basic knowledge in either C++ or Python
- Interest to work with surgeons
- Interest to work in a scientific manner
- Basic knowledge of X-ray and CT modality
- Basic knowledge of projective geometry
- Prior experience with 3D data
Literature
[1] El-Azab, H. M., Morgenstern, M., Ahrens, P., Schuster, T., Imhoff, A. B., & Lorenz, S. G. (2011). Limb alignment after open-wedge high tibial osteotomy and its effect on the clinical outcome. Orthopedics, 34(10), e622e628. https://doi.org/10.3928/01477447-20110826-02 [2] Wright, John M. MD; Crockett, Heber C. MD; Slawski, Daniel P. MD; Madsen, Mike W. MD; Windsor, Russell E. MD High Tibial Osteotomy, Journal of the American Academy of Orthopaedic Surgeons: July 2005 - Volume 13 - Issue 4 - p 279-289 [3] Lee DC, Byun SJ. High tibial osteotomy. Knee Surg Relat Res. 2012;24(2):61-69. doi:10.5792/ksrr.2012.24.2.61 [4] Li, S., Xu, C., & Xie, M. (2012). A robust O (n) solution to the perspective-n-point problem. IEEE transactions on pattern analysis and machine intelligence, 34(7), 1444-1450. [5] Markelj, P., Tomaevič, D., Likar, B., & Pernu, F. (2012). A review of 3D/2D registration methods for image-guided interventions. Medical image analysis, 16(3), 642-661. [6] Esteban, J., Grimm, M., Unberath, M., Zahnd, G., & Navab, N. (2019, October). Towards fully automatic X-ray to CT registration. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 631-639). Springer, Cham. [7] Grupp, R. B., Unberath, M., Gao, C., Hegeman, R. A., Murphy, R. J., Alexander, C. P., ... & Taylor, R. H. (2020). Automatic annotation of hip anatomy in fluoroscopy for robust and efficient 2D/3D registration. International Journal of Computer Assisted Radiology and Surgery, 1-11. [8] Unberath, M., Zaech, J. N., Gao, C., Bier, B., Goldmann, F., Lee, S. C., ... & Navab, N. (2019). Enabling machine learning in x-ray-based procedures via realistic simulation of image formation. International journal of computer assisted radiology and surgery, 14(9), 1517-1528. [9] Grimm, M., Esteban, J., Unberath, M., & Navab, N. (2020). Pose-dependent weights and Domain Randomization for fully automatic X-ray to CT Registration.| Students.ProjectForm | |
|---|---|
| Title: | Intraoperative 2D-3D Registration for Knee Alignment Surgery |
| Abstract: | In this project a system is developed, which assists surgeons in verifying a surgical result by comparing interventional X-Rays to a preoperative plan. The targeted surgical procedure is knee osteotomy in which a bone (usually the tibia) is cut at a specific point and the two segments are repositioned to correct knee alignment. The 2D X-Ray images are to be registered to the 3D preoperative plan in order to compute the achieved 3D configuration between the two bone segments. This allows the surgeon to make sure that the plan is carried out accurately, or make adjustments if necessary, as correct knee alignment leads to improved patient outcome. |
| Student: | |
| Director: | Prof. Dr. Nassir Navab |
| Supervisor: | Alexander Winkler, Matthias Grimm |
| Type: | DA/MA/BA |
| Area: | |
| Status: | running |
| Start: | |
| Finish: | |
| Thesis (optional): | |
| Picture: | |