|
M. Müller, M. Yigitsoy, H. Heibel, N. Navab
Deformable Reconstruction of Histology Sections using Structural Probability Maps
International Conference on Medical Image Computing and Computer Assisted Interventions (MICCAI), Boston, USA, September 2014
(bib)
|
|
T. Peng, M. Yigitsoy, A. Eslami, C. Bayer, N. Navab
Deformable registration of multi-modal microscopic images using a pyramidal interactive registration- learning methodology
6th Workshop on Biomedical Image Registration, London, UK, July, 2014
(bib)
|
| 2013 |

|
M. Yigitsoy, N. Navab
Structure Propagation for Image Registration
IEEE Trans. Med. Imag., vol. 32, no. 9, pp. 1657-1670, September 2013
(bib)
|
Team
Contact Person(s)
Working Group
Location
 Visit our lab at Garching.
internal project page
Visit our lab at Garching.
internal project page
Please contact
Olivier Pauly for available student projects within this research project. -->
Student Projects
We have the following open master thesis. If you are interested in any of them, please contact to either
Olivier Pauly or
Mehmet Yigitsoy. See the descriptions of each project below.
- (Open) Mitochondria Detection and Tracking in Microscopic Images, in collaboration with Thomas Misgeld Lab, Institute of Neuroscience, TUM
- (Open) Motion Compensation in in-vivo 3D Microscopic Data, in collaboration with Thomas Misgeld Lab, Institute of Neuroscience, TUM
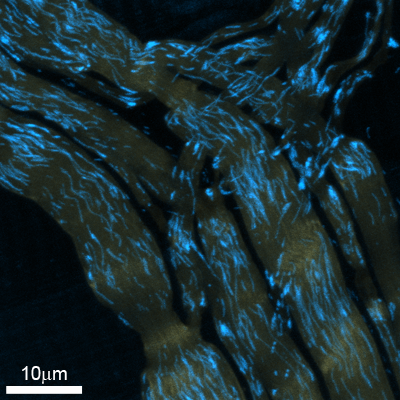
Mitochondria Detection and Tracking in Microscopic Images
|
Abstract:
Nerve cells – due to the length of their processes – are highly dependent on organelle transport and their cytoskeletal support scaffold. New approaches now allow visualizing moving organelles and cytoskeletal dynamics in vivo or in acutely explanted tissue (Misgeld et al., Nature Methods 2007; Marinković et al., PNAS, in press). For this, transgenic mice are used, in which fluorescent proteins are targeted into organelles or fused to cytoskeletal building blocks. Time-lapse microscopy of such mice (or tissues derived from them) can generate a deluge of data, in which particles need to be tracked, often under demanding optical conditions (e.g. as particles move behind or across others that are similarly labeled; or because the signal characteristics change locally as the track crosses scattering or fluorescent objects above or below the plane of focus).
Developing algorithms to faithfully track particles under such conditions would allow dramatic “up-scaling” of tracking experiments. As axonal transport and cytoskeletal stability are central targets of many neurodegenerative changes (probably including Alzheimer’s disease and motor neuron degeneration), testing pharmacological or genetic modulators of particle behavior is important. The development proposed for this project would make a seminal contribution in this important area of disease-related research.
|
The Goals:
- Development of an approach for detecting mitochondria
- Development of a tracking approach
- Quantitative analysis of the mitochondria transport
Required Skills:
- Good programming skills in C++, Matlab and other languages
- Interest in working in interdisciplinary projects
- Knowledge in machine learning and tracking
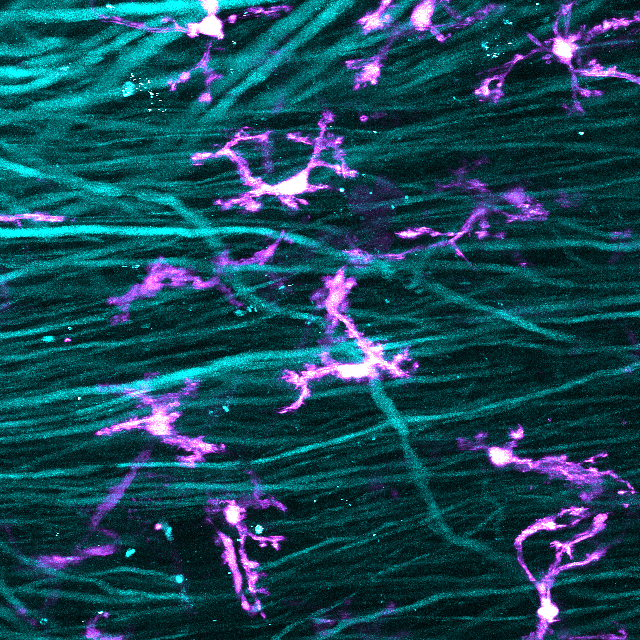
Motion Compensation in in-vivo 3D Microscopic Data
|
Abstract:
Recent advances in fluorescence microscopy have opened up the ability to probe deep into living tissue. However, movement artifact imposes a large barrier to successful 3D reconstruction of in vivo microscopy data. Although there are some whole animal preparations in which a tissue of interest can be held completely immobile while maintaining the viability of the sample, these preparations are limited in the range of anatomical structures that can be accessed. This limitation makes the in vivo study of many anatomical structures difficult or near impossible in three dimensional space.
A universal solution to in vivo movement artifact that avoids difficult custom designed stabilization procedures would be a large step forward for in vivo microscopy studies. Removing the movement artifact using post hoc adjustment of primary microscopy data is an ideal approach. Stretching algorithms have already been applied to two dimensional time lapse data and made possible the tracking of cellular activity across time. However, it is common to simply throw out z-shifted data points in such experiments. Similar procedures would obviously not be applicable for three dimensional reconstructions.
In vivo multi-photon microscopy time-lapse datasets from mouse spinal cord imaging experiments will be used for this project (Kerschensteiner et al., Nature Medicine 2005; Nikić et al., Nature Medicine 2011). Even though the mouse is held relatively stable in spinal clamps during image acquisition, movement artifacts from respiration and in some cases heart-beat are observed. Distortions can manifest in all dimensions but are most prominent along the z-axis. Importantly, the movement artifact is not constant across samples and depends heavily on the experimental procedures as well as the status of anesthesia. In vivo samples can be referenced to a post hoc image stack after the mouse is killed and the sample is totally stable. While the immediate goal of the project is to find a solution for movement artifact of in vivo spinal cord data, ideally this project would be a starting point for a universally applicable solution to movement artifact in in vivo fluorescence microscopy.
|
The Goals:
- Development of a deformable registration approach
- Development of a motion compensation approach
Required Skills:
- Good programming skills in C++, Matlab and other languages
- Interest in working in interdisciplinary projects
- Prior knowledge and interest in medical image registration




