Stain Separation and Structure-Preserving Color Normalization for Histological ImagesScientific Director: Nassir NavabContact Person(s): Tingying Peng |
Abstract
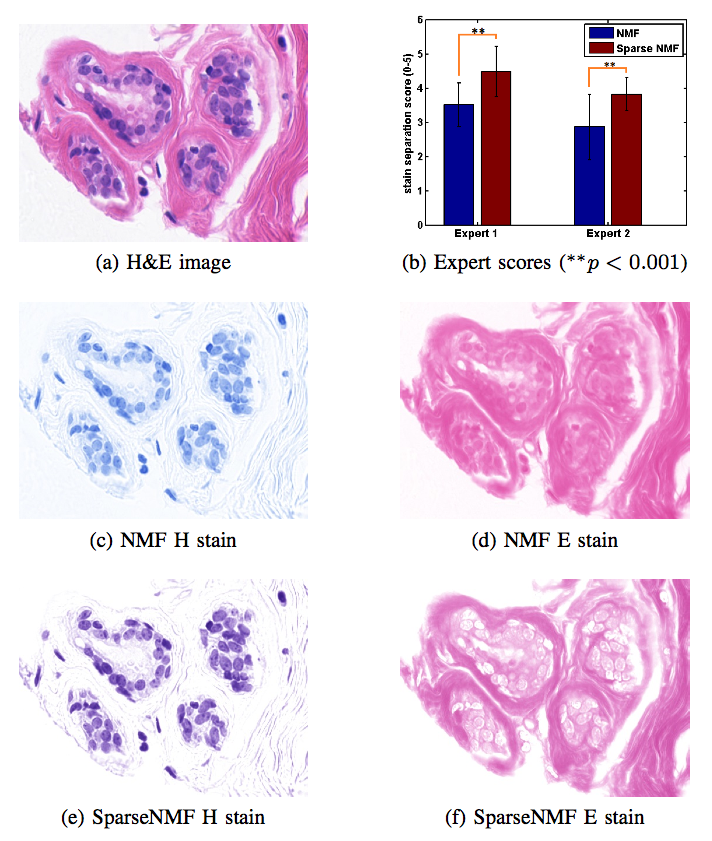
Staining and scanning of tissue samples for microscopic examination is fraught with unwanted variations that affect their color appearance. Sources of these variations include differences in raw material and manufacturing techniques of stain vendors, staining protocols of labs, and color responses of digital scanners. Color normalization of stained biopsies and tissue microarrays will help pathologists and computational pathology software while comparing different tissue samples. However, techniques that are used for natural images, such as histogram matching fail to utilize unique properties of stained tissue samples and produce undesirable artifacts. Tissue samples are stained with only a few reagents (frequently only two -- hemotoxylin and eosin or H\&E) and most tissue regions bind to only one stain or the other, thus producing sparse density maps composed of only a few components. This underlying structure of sparse stain density is biomedically important. We used these properties of stained tissue to propose a technique for stain separation and color normalization. Based on sparse non-negative matrix factorization (sparseNMF), we estimate prototype color and density map of each stain in an unsupervised manner to perform stain separation. To color normalize a given source image, we combine its stain density maps with the stain color prototypes of a target image whose appearance was preferred by pathologists. In this way, the normalized image preserve the biological structure encoded in the stain density of the source image. Both the proposed sparseNMF stain separation and color-normalization techniques yield higher correlation with ground truth than the state of the art. They are also rated qualitatively higher than other techniques by a group of pathologists. We further propose a computationally faster extension of this technique for large whole-slide images that selects an appropriately small sample of patches to compute the color prototypes of each stain instead of using the entire image. The fast scheme achieves a 20-folds acceleration, which does not only greatly enhance the analysis efficiency, but also allow its clinical applications to become practically feasible.Team
Contact Person(s)
|
Working Group
|
|
|
|
|
Location
| Technische Universität München Institut für Informatik / I16 Boltzmannstr. 3 85748 Garching bei München Tel.: +49 89 289-17058 Fax: +49 89 289-17059 |
| Klinikum rechts der Isar der Technischen Universitüt München Ismaninger Str. 22 81675 München IFL Lab - Room: 01.3a-c Tel.: +49 89 4140-6457 Fax: +49 89 4140-6458 |
internal project page
Please contact Tingying Peng for available student projects within this research project.